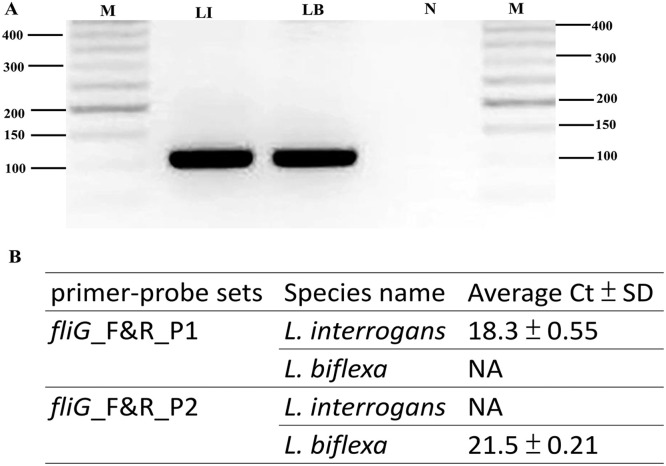
Fig. 4.
Validation of novel primer-probe sets for Leptospira species detection. (A) PCR amplification was performed using the primer pair fliG_F and fliG_R. Each reaction contained 25 ng of Leptospira genomic DNA under standard conditions. The amplification assay consisted of 30 cycles of 95 °C for 15 s, 60 °C for 75 s, and 72 °C for 75 s. Subsequently, the samples were maintained at 72 °C for 10 min, and the PCR products were separated via agarose gel electrophoresis and photographed. The genomic DNA from L. interrogans yielded a 110 bp fragment (Lane LI); genomic DNA from L. biflexa amplified a 110 bp fragment (Lane LB); negative control (water; Lane N); M, molecular size determined using a 50-bp ladder molecular marker. (B) RT‒PCR amplification was performed using the primer pairs fliG_F and fliG_R combined with different probes designed in this study. Duplicate Ct values were averaged, and the standard deviation (SD) of the Ct values was calculated. NA: samples with no amplification detected.