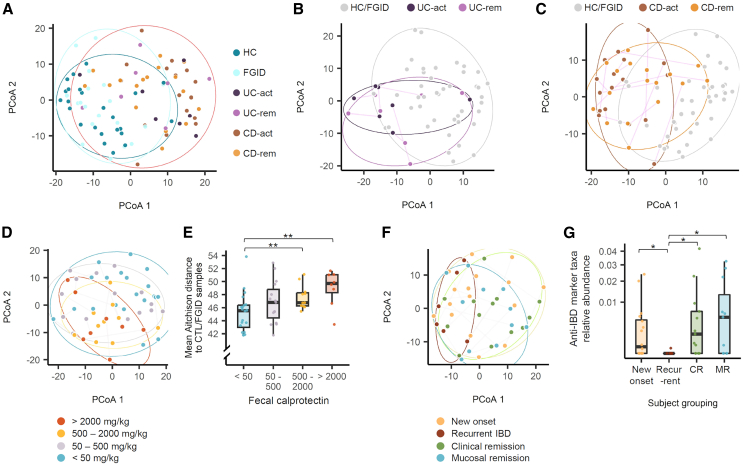
Figure 3.
Gut bacterial community variation associated with patients’ clinical features
(A) PCoA of gut microbiota using read counts per 16S ASV. Aitchison distance was used to quantify inter-sample differences. Adonis 2 test: UC-act vs. HC and FGID, R2 = 0.046, p = 0.001; CD-act vs. HC and FGID, R2 = 0.078, p = 0.001.
(B and C) Individual trajectories within the patients with UC (B) and CD (C) over two sampling time points. Adonis 2 test: UC-act vs. UC-rem, R2 = 0.069, p = 0.63; CD-act vs. CD-rem, R2 = 0.038, p = 0.078.
(D and E) Comparison of fecal samples based on fecal calprotectin levels using PCoA (D) and mean distance to healthy controls (E). Adonis 2 test: low vs. high, R2 = 0.052, p = 0.01; low vs. very high, R2 = 0.076, p = 0.001.
(F and G) Comparison of new-onset patients, those with recurrent active IBD, those in CR, and those in MR. In (A–D) and (F), ellipses are drawn around each sample group using the geom_mark_ellipse function of the ggforce package, and statistical significance of inter-group variation was tested by Adonis2 permutational multivariate analysis of variance implemented in the vegan package based on Aitchison distance. In (A), the patients with IBD (UC-act, UC-rem, CD-act, and CD-rem) are circumscribed together in a single ellipse. In (E) and (G), the boxplots represent interquartile range, along with the individual data points shown as dots. The pairs of groups with values of p < 0.05 as determined by the Wilcoxon test are indicated by horizontal brackets: ∗p < 0.05, ∗∗p < 0.01. ASV, amplicon sequence variant; CD, Crohn disease; CD-act, active CD; CD-rem, remission CD; CR, clinical remission; MR, mucosal remission; PCoA, principal coordinate analysis; UC, ulcerative colitis; UC-act, active UC; UC-rem, remission UC.