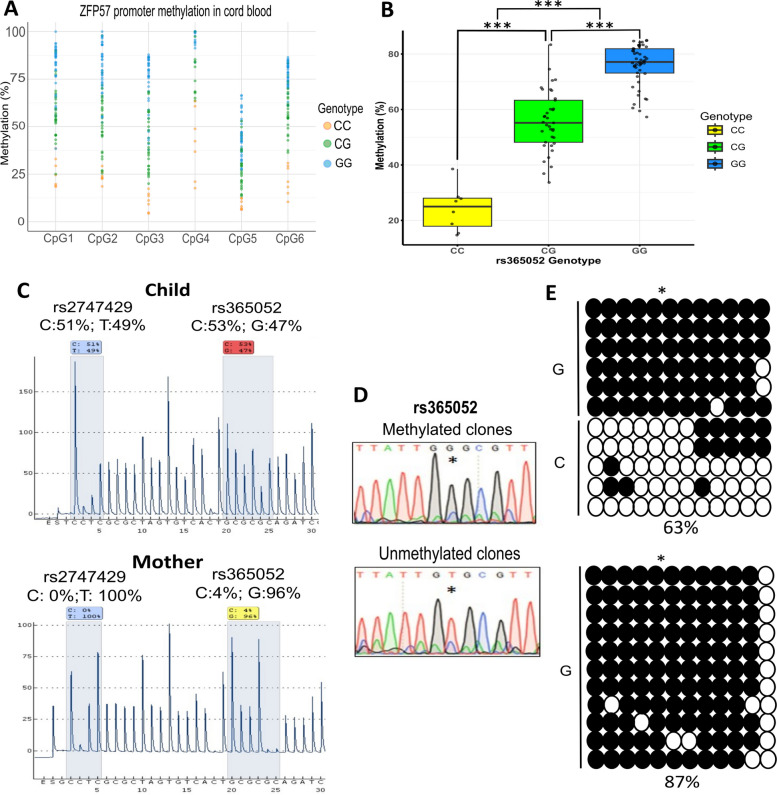
Fig. 3.
Methylation quantitative trait loci in the differentially methylated region of ZFP57. A Detailed CpG methylation analysis of the ZFP57 alternative promoter analysed by pyrosequencing in the cord blood. DNA methylation of individual CpG sites is shown for CpG sites 1–6, using primers in Additional File 1: Table S1 (CpG pyroassay). Yellow dots represent samples with C/C genotype within the rs365052 SNP, green represent samples with C/G and blue with a G/G SNP genotype. B Boxplot with jitter data of ZFP57 methylation by the rs365052 SNP demonstrating significant methylation differences between different genotype groups in blood. Genotype analysis performed by allele quantification (AQ). Statistically significant values are marked as: *** indicates p < 0.001, conducted by ANOVA. C Identification of heterozygous gDNA sample (child) for the rs365052 (C/G) region assessed by the AQ assay (SNP pyroassay 1). Allele frequency of 2 SNP variants rs2747429 (C/T) and rs365052 (C/G) indicated by the common SNPs analysis. Percentage of the allele frequency is highlighted in boxes with colour identification based on the quality of the run (blue—very good, yellow—satisfactory, red—poor quality). AQ assay of matching homozygous maternal gDNA within the SNP variant rs365052 (G/G) extracted form blood. D Sanger sequencing analysis identifying mQTL within the rs365052 SNP that in methylated and unmethylated clones, SNP shown at asterisk. E Bisulfite sequencing clonal analysis of the region from Fig. 2 for child (C/G) and mother (G/G), showing methylation distribution in individual clones. Polymorphic base located between the fifth and sixth CpG site (asterisk) corresponds with the rs365052. Full circle represents methylated and empty unmethylated CpG sites