Abstract
Fragile X Syndrome (FXS) is a neurodevelopment disorder characterized by cognitive impairment, behavioral challenges, and synaptic abnormalities, with a genetic basis linked to a mutation in the FMR1 (Fragile X Messenger Ribonucleoprotein 1) gene that results in a deficiency or absence of its protein product, Fragile X Messenger Ribonucleoprotein (FMRP). In recent years, mass spectrometry (MS) – based proteomics has emerged as a powerful tool to uncover the complex molecular landscape underlying FXS. This review provides a comprehensive overview of the proteomics studies focused on FXS, summarizing key findings with an emphasis on dysregulated proteins associated with FXS. These proteins span a wide range of cellular functions including, but not limited to, synaptic plasticity, RNA translation, and mitochondrial function. The work conducted in these proteomic studies provides a more holistic understanding to the molecular pathways involved in FXS and considerably enhances our knowledge into the synaptic dysfunction seen in FXS.
Keywords: Fragile X syndrome (FXS), Fragile X messenger ribonucleoprotein (FMRP), Proteomics, Mass spectrometry, Neurodevelopment disorders
1. Introduction
Representing one of the most common forms of monogenic inherited intellectual disability, fragile x syndrome (FXS) has an incidence of 1: 2500–5000 in males and 1: 4000–6000 in females (Berry-Kravis, 2014). FXS is caused by a CGG trinucleotide repeat expansion mutation in the promoter region of the FMR1 (Fragile X Messenger Ribonucleoprotein 1) gene (Lozano et al., 2014). The CGG repeat sequence is transcribed into the FMR1 mRNA, but since this CGG repeat is located in the 5′ untranslated region (UTR), the length of the expansion does not affect the sequence of its protein product, Fragile X Messenger Ribonucleoprotein (FMRP) (Berry-Kravis, 2014). Normal alleles, characterized by 5–40 CGG repeats, are typically stable on transmission, but certain alleles at the upper end of this range (particularly those with no AGG interruptions in the CGG sequence) can become unstable and expand during maternal transmission giving rise to gray zone (41–54 CGG repeats) alleles or with further expansion, premutation alleles (55–200 CGG repeats) (Salcedo-Arellano et al., 2020; Telias, 2019). Although premutation alleles do not cause the FXS phenotype, they are vulnerable to large increases in repeat length during meiosis, and to expansion to over 200 repeats during female meiosis. Therefore, unlike the typical pattern of inheritance for an X-linked disease, in order for FXS to manifest, the premutation must be passed down and expanded enough to reach a critical length (>200 repeats) (Santoro et al., 2012). Upon the expansion reaching >200 CGG repeats, de novo methylation, hypoacetylation of associated histones, and heterochromatin formation are induced causing transcriptional silencing of FMR1, leading to a deficiency or absence of FMRP (Bagni et al., 2012; Santoro et al., 2012) (Fig. 1).
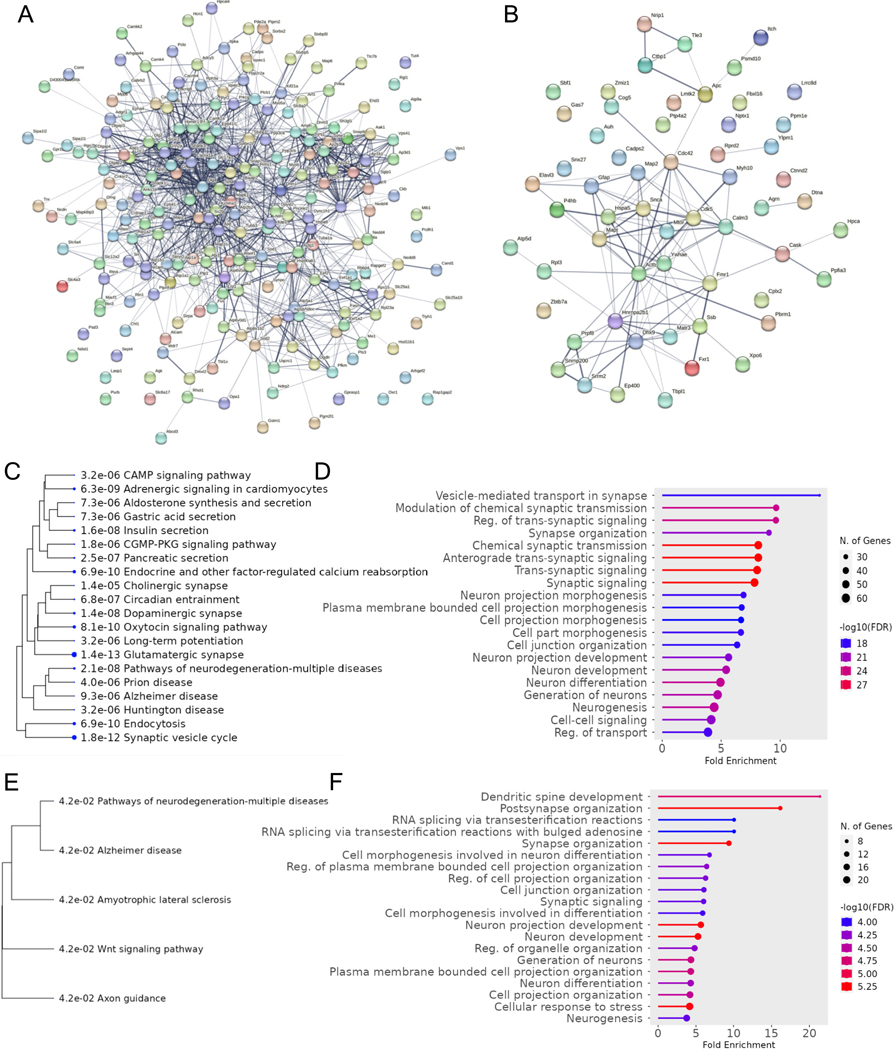
Fig. 1. Bioinformatic Analysis of Differential Cortical Proteins Translationally Regulated by FMRP.
A) Protein-protein interaction network of upregulated cortical proteins. B) Protein-protein interaction network for downregulated cortical proteins. C) Pathway enrichment for upregulated cortical proteins. D) GO enrichment for upregulated cortical proteins. E) Pathway enrichment for downregulated cortical proteins. F) GO enrichment for downregulated cortical proteins. Analysis was done in ShinyGo 0.80 and String (Ge et al., 2020).
The study of FXS has revealed many pathways that are crucial to learning and memory formation and much of this knowledge has been applied to understanding broader non-Mendelian forms of intellectual disability, such as autism spectrum disorder (Woods et al., 2015). Since the FMR1 gene was identified as the gene mutated in FXS in 1991 (Verkerk et al., 1991), there has been great progress in understanding the genetics of FXS and FMRP function, and how FMRP loss leads to the many aberrant cellular, molecular, and behavioral observations seen in FXS. FMRP is a brain enriched RNA-binding protein that is associated with regulating the translation of mRNAs responsible for proper neuronal development (Sethna et al., 2014). This is demonstrated by the observation that the loss of FMRP in FXS adversely impacts many aspects of brain development and cellular function including neurogenesis, dendritic morphology, synaptogenesis, and circuitry integration. These observations have prompted studies directed towards identifying FMRP target mRNAs using high resolution techniques such as CLIP-seq, demonstrating the role of FMRP in regulating the translation of mRNAs involved many critical brain pathways such as, but limited to, glutamate receptor signaling (e.g., GRIN2A/2B, GRIK3); synaptic long term depression and potentiation pathways (e.g., MAPK1, CAMK2A/2B, CREBBP); GABA receptor signaling (e.g., GABAR1/R2) (Darnell et al., 2011); cAMP/cGMP signaling (e.g., PDE2A) (Maurin et al., 2018); regulation of RAC1 signaling; and pathways involved in circadian expression (e.g., Npas2, Ppargc1a, Ncoa2) (Sawicka et al., 2019). Many neuronal targets for treating FXS have been identified through these studies, however; questions central to the development of therapeutics and treatment for individuals with FXS remain unsolved (Berry-Kravis, 2014; Berry-Kravis et al., 2011).
Although initial research efforts on FXS were highly genomics and transcriptomics driven, in recent years, proteomic studies of FXS, especially those utilizing mass spectrometry (MS), have gained momentum as the field strives to identify protein markers for therapeutic intervention; however, these studies are still very limited and should be further investigated for many reasons. Proteomics complements genomic and transcriptomic information since dynamics at the developmental and genetic stages are reflected at the protein level (Ngounou Wetie et al., 2014). Furthermore, MS can reveal protein expression levels, protein interaction networks, and important post-translation modifications at the protein level, making it potentially more closely related to mechanisms underlying human disorders in comparison with molecular genetics (Pal et al., 2015; Wormwood et al., 2019). Since proteins are active effectors of biological processes, protein biomarkers can be used as therapeutic targets or in a preclinical setting to measure the efficacy of treatment trials.
In the context of linking the molecular biology of FMRP to FXS, MS holds great potential to enhance our knowledge on FMRP function since the effects of FMRP loss should be measured at the protein level. Early models of FMRP-dependent translation regulation support that FMRP acts as a translational repressor, and in the absence of FMRP translation is increased. While this holds true, this model is too simplistic and has been enhanced though recently published literature. This model was supported by the HITS-CLIP experiment of Darnell et al., which revealed that an increased number of ribosomes are stalled on FMRP-targeted transcripts, and this stalling event occurred on coding sequences of target mRNAs (Darnell et al., 2011). FMRP has also been shown to repress translation through interacting with translational machinery, in the miRNA pathway through its association with the RISC complex, and may participate in the transport of its target mRNAs along dendrites within ribonucleoprotein complexes/transport granules to repress translation until they arrive at the synapse (Richter and Zhao, 2021). However, more recently published studies reveal divergence in the translational repression model and add to the already complex functional nature of FMRP. Greenblatt and Spradling et al., analyzed Drosophila oocytes on the basis that this model depends heavily on translating stored mRNA in a similar fashion like neural synapses. Their ribosomal profiling data revealed that FMRP acts preferentially on large proteins to enhance their translation, in which many of these transcripts are autism-related transcripts (Greenblatt and Spradling, 2018). A different study conducting FMRP cTag CLIP on hippocampal CA1 pyramidal neurons supported this finding and demonstrated that the decrease in mRNA translation as a result of FMRP loss is directly proportional to the amount of FMRP binding, suggesting FMRP may stabilize its target mRNAs or acts to protect mRNAs from degradation (Sawicka et al., 2019). This study also highlights the cell-type specificity of FMRP-targeted mRNAs through revealing 246 transcripts that uniquely identified in CA1 neurons versus transcripts in cerebellar excitatory neurons (Sawicka et al., 2019), which was also demonstrated in a seperate CLIP-seq study aimed to identify FMRP targets in human dorsal and ventral neural progenitor cells and neurons (Li et al., 2020). Multiple studies corroborate that FMRP preferentially binds longer mRNAs (Hale et al., 2021; Li et al., 2020; Sawicka et al., 2019; Seo et al., 2022); however, why FMRP preferably targets longer mRNA transcripts is not fully understood. The multiomic study of Seo et al., which involved proteomics and Translating Ribosome Affinity Purification and RNA-seq (TRAP-seq) provides insight to this model and demonstrates how MS complements transcriptomic data in elucidating FMRP’s function (Seo et al., 2022). Here, they observed an increase in translation of ribosomal mRNAs, which also manifested in an overexpression of ribosomal proteins in Fmr11/y CA1 neurons. Their TRAP-seq data also demonstrated that Fmr11/y neurons exhibit a length dependent imbalance in mRNA translation where shorter transcripts are favored versus longer transcripts, which would provide evidence to why shorter transcripts such as ribosome and mitochondrial transcripts are enhanced whereas longer synaptic transcripts are repressed (Seo et al., 2022). This size effect was also reflected at the protein level as revealed by their proteomics data and suggests that overexpression of ribosomal proteins may exacerbate translation of shorter transcripts versus longer transcripts since shorter transcripts have higher translation efficiency (Seo et al., 2022). Notably, this profile was also observed when stimulating mGluR-LTD in WT CA1 neurons and treatment with CX-5461, a ribogenesis inhibitor, rescued this length-dependent shift in translation and inhibits mGluR-LTD, pointing towards therapeutic avenues targeting ribogenesis for FXS (Seo et al., 2022). In addition to the cell type specificity of FMRP targets, a recent study supports the model that subcellular location of FMRP and FMRP-targeted mRNAs facilitates FMRP to regulate the expression of different targets within different compartments in a single neuronal cell type. This study revealed an enrichment of mRNAs encoding nuclear proteins in the cell body of CA1 neurons whereas an enrichment of mRNAs encoding synaptic proteins was found in the dendrites of CA1 neurons (Hale et al., 2021). These studies strongly suggest that FMRP has distinct roles in different cell types and within different cellular compartments, which could suggest proteome changes in models of FXS may also exhibit similar characteristics, necessitating the need to investigate cell-type specific proteomes and subproteomes within the same cell types.
Although mRNA target selection helps define the function of FMRP, multiple studies, including some studies of FXS or animal model tissues, have demonstrated that global mRNA expression can be positively, but weakly, correlated to protein expression (Bowling et al., 2019; Liao et al., 2008; Macklin et al., 2020; Tang et al., 2015). This discord is likely due to the highly complex and dynamic nature of proteome regulation since protein expression is affected by processes such as alternative splicing, transcript and protein degradation, protein-protein interactions, and post translational modifications. Although transcriptomic studies have been valuable to the FXS field as they paved a trajectory for studying disease relevant mechanisms, they are not able to capture the secondary effects of FMRP loss on the proteome. Since FMRP has direct and indirect modes of regulating translation and promoting expression of its target mRNAs, we cannot be certain that direct mRNA targets of FMRP are dysregulated in FXS. These changes can only be reliably captured by direct quantitation of the proteome, which further supports the implementation of complementary proteomics research for FXS. This is not to say that the field has overlooked the potential of proteomic studies to investigate FXS, but rather reflects that many researchers might not have access to the instrumentation needed to carry out these high throughput experiments. Even with this limitation in place, there have been a handful of studies published utilizing MS as a tool for studying FXS (See Tables 1 and 2). The purpose of this review is to provide a comprehensive literature summary on how MS-based proteomics has been applied to the study FXS and provide insights into future opportunities.
Table 1.
Summary of Proteomics Studies Using Animal Models of FXS.
| Reference | Experimental Design | Key Findings |
|---|---|---|
| Liao et al., 2008 | Synaptosome fractions from WT and Fmr1 cortical neurons (n = 18) analyzed by SILAC- based quantitative LC-MS | • Increased expression of proteins involved in synaptic pruning, synaptic formation, and synaptic maturation in Fmr1 KO mice (e.g., APC, FUS, tPA, SERBP, and N-Cam) • Decreased expression of Kcnma1 in Fmr1 KO mice |
| Klemmer et al., 2011 | Hippocampal synapses from postnatal day-14 WT and Fmr1 KO mice (n = 8 each group) analyzed by iTRAQ-based quantitative LC/MS | • Upregulation of proteins involved in cell differentiation and neurite outgrowth (e.g., Basp1, Gap43, and Cend1) and synaptic vesical release (e.g., syntaxin-1B2, synaptsin-1) in Fmr1 KO mice |
| Broek et al., 2016 | Analysis of synaptosome isolates from 14-week-old Fmr1 KO (n = 12) and WT (n = 11) by label-free LC-MS and target validation by SRM-MS | • Altered expression of proteins involved in GABAergic/glutaminergic transmission and ATPases showed inverse expression levels in hippocampal (downregulated) and cerebellar (upregulated) regions |
| Tang et al., 2015 | Examination of age-dependent alterations in neocortical synaptosome-enriched samples from postnatal day-14 (n = 4) and postnatal day-45 (n = 4) Fmr1 KO mice against age-matched WT mice (n = 4 each group) using SILAM and high-resolution MS | • Higher number of differential proteins between day-17 Fmr1 KO and WT mice compared to day-45 Fmr1 KO and WT mice. • Upregulation of core presynaptic vesicle proteins (e.g., synapsin 1, Snap25), postsynaptic channels (e.g., Hcn1), synaptic scaffolding proteins (e.g., Shank family proteins, Psd95) • Downregulation of postsynaptic receptor mGluR7 |
| Bowling et al., 2019 | BONLAC proteomics and SILAC labeling on 10–12-week-old Fmr1 KO and WT mice (n = 9 each group) to compare difference in translation during steady state and in response to mGluR5 signaling | • 324 altered newly synthesized proteins at steady state enriched in proteins involved in biosynthesis, protein-folding, and synaptic proteins (e.g., upregulation in SynGAP and mGluR5; downregulation in NMDAR2B) • 278 altered newly synthesized proteins in response to DHPG stimulation enriched with cell membrane and biosynthesis proteins (e.g., downregulation of Ras and Hk1) |
| Bülow et al., 2021 | Analysis of steady state proteome changes after chronic activity deprivation in WT and Fmr1 KO cortical neurons from P0-P1 mice using 22-plex TMT-based quantitative LC-MS | • 260 altered proteins in Fmr1 KO cells after drug treatment • Proteins sensitive to activity blockade localized to the mitochondrial membrane and play roles in the TCA cycle and electron transport chain (e.g., Ndufa11, Atp5d) |
| Kuzniewska et al., 2020 | iTRAQ and TMT-based quantitative LC-MS and in vitro stimulation of isolated mouse synaptosomes in 1–2-month-old Fmr1 KO and age-matched WT mice (n = 4 each group) | • 516 proteins found significantly altered in Fmr1 KO mice, specifically, 69 locally synthesized proteins at the synapse were mitochondrial proteins incorporated into respiratory chain complexes. • Fmr1 KO mitochondria show elevated levels of respiration |
| Licznerski et al., 2020 | MS analysis of Fmr1 KO neurons from P1-P2 mice | • Increased expression in glycolic enzymes such has hexokinase 1/2, pyruvate kinase, and lactate dehydrogenase. • Neurons exhibit mitochondrial inner membrane leak |
| Ma et al., 2024 | MALDI-MS imaging and LC-MS analysis on hippocampal and hypothalamic regions in Fmr1 KO mice on soy-based diets | • Soy consumption in mice is associated with mitochondrial metabolism proteins such as glycolic and TCA enzymes |
| Talvio et al., 2021 | ICP-MS analysis to investigate trace elements in Fmr1 KO mice | • Increased iron content in the heart and cerebellum in Fmr1 KO mice |
| Ceman et al., 2003 | Analysis of mouse fibroblast L- M(TK-) cells and mouse brain lysates using MALDI-TOF MS, and site directed mutagenesis | • Revealed phosphorylation site of FMRP at S499 • Phosphorylation of FMRP influences translation state of FMRP associated polyribosomes |
| Matic et al., 2014 | Quantitative analysis of proteome and phosphoproteome of Fmr1 KO MEFs using MS and SILAC | • Decreased expression of Prp and cPLAC2 in Fmr1 KO mice • Dysregulation of proteins and phosphorylation events in major signal transduction pathways (e.g., MEK/ERK pathways was downregulated with decreased phosphorylation of Erk1/2, IRIS, and Rictor and decreased protein expression ofPTEN) |
| Wu et al., 2022 | 6-plex TMT quantitative proteomics on peptides enriched by anti-K-ac antibodies to analyze the lysine acetylome in the hippocampus of Fmr1 KO mice | • 51-K-ac peptides localized to 45 proteins were significant altered • Enriched for proteins involved in carboxylic acid metabolism, aerobic respiration, and TCA enzymes (e.g., upregulation of DLD, downregulation of PDHA1) |
| Zhang et al., 2005 | Analysis of brain proteome of 2-day-old dfmr1 null mutants against WT controls (n = 75 each group) using DIGE and MALDI-TOF MS | • Altered expression in multiple isoforms of proteins involved in monoamine synthesis, phenylalanine hydroxylase and GTP cyclohydrolase 1. • Elevation of cytoskeleton proteins β-tubulin and actin 5C |
| Monzo et al., 2010 | Analysis of dfmr1 mutant embryos using 2D DIGE/ MALDI-TOF MS | • 3 subunits of the chaperonin containing TCP-1 complex were misregulated in Dfmr1 mutants |
| Xu et al., 2018 | TMT quantitative proteomics analysis of hippocampus, cortex, and testis in Fmr1 KO mice against WT mice (n = 4) each group | • Downregulation of ribosomal protein subunits in the hippocampus and cortex of Fmr1 mice • Neuroplasticity related protein, Ly-6/neurotoxin like protein 1 found to be upregulated in the cortex but not the hippocampus in Fmr1 KO mice. • Neuromodulin and nestin were upregulated in hippocampus and downregulated in the cortex of Fmr1 KO mice |
| Kieffer et al., 2022 | LC-MS/MS analysis of affinity pull downs using rat forebrain nuclei enriched fractions and GST fusion protein fragments of N-terminal domains of FMRP to identify FMRP protein interactors | • Identification of three novel FMRP-interacting proteins Ddx4, Poldip3, Hnrnpa3 • FXRIP, a known FMRP interactor in the cytoplasm, also binds FMRP in the nucleus |
Table 2.
Summary of Proteomics Studies Using Human Models of FXS and Human Tissue Samples from Individuals with FXS.
| Reference | Experimental Design | Key Findings |
|---|---|---|
| Godler et al., 2010 | MALDI-TOF MS for FMR1 locus methylation mapping from blood samples from controls (n = 49), gray zone carriers (n = 18), premutation carriers (n = 22) and FXS subjects (n = 22) | • Positive correlation between methylation status of FREE1/FREE2 in high functioning males, including FXS individuals with mosaicism. • Negative correlation between FREE1/FREE2 and number of FMRP-positive lymphocytes and FMR1 activation ratio in blood of high functioning males and females |
| Godler et al., 2012 | Blood analysis on controls (n = 72), premutation carriers (n = 62) and full mutation females (n = 18) with previously conducted Wechsler IQ tests using MALDI-TOF MS | • Methylation patterns on FREE2 region showed methylation of intron 1 CpG sites 10–12 showed highest diagnostic sensitivity and specificity for detecting females with standardized verbal IQ of <70. • In full mutation individuals, methylation pattern of the same sites correlated with full-scale, verbal, and performance IQ |
| Zhang et al., 2019 | Expression and characterization of human FMRP isoforms and interacting proteins in human HEK293 cells by LC-MS/MS | • Peptides SFLEFAEDVIQVPR and MEELVVEVR represent the most sensitive peptides for targeted quantification analysis of FMRP. • BCLAF1, NUFIP2, and FXR1 as protein interactors of FMRP |
| Taha et al., 2021 | LC-MS/MS analysis of affinity pull downs using HeLa cells and GST fusion protein fragments of N-, C-, and central domains of FMRP to identify FMRP protein interactors | • Stress granule proteins, NONO and G3BP1, were found to be FMRP interactors. • FMRP and G3BP1 colocalize in stress granules |
| Shen et al., 2023 | Co-IP-MS analysis of human cortical neurons differentiated from FMRP-FLAG hPSCs | • Critical role of species-specific FMRP regulation of RACK1 in prenatal cortical development |
| Utami et al., 2020 | Transcriptomic and proteomic analysis on day-37 FMR1 KO, FXS, and control neurons (n = 4 each group) | • differential proteins in the FXS neuron showed enrichment in vesicle transport and synaptic signaling, cell cycle processes, DNA replication, and DNA metabolism. • Downregulation of CNTNAP2, GPRIN3, KIF5c, and CNTN1 in FXS neurons |
| Kurosaki et al., 2022 | Integrative omics analysis of human FMR1 KO SH-SY5Y neuroblastoma cells using SILAC and MS | • FMR1 cells undergo largest increase in translation, destabilizing, and decay. • FMRP stabilizes mRNAs and decreases translation by binding solely to the 5’UTR, coding sequence, or 3’UTR via GC rich sequences. • Increase abundance in CHD5, UNC13D, RPH3A in FMR1 KO cells. • Decrease abundance in MED1 and PGAP1 FMR1 KO cells |
| Dionne and Corbin, 2021 | Analysis of the nascent proteome in PBMCs from male FXS patients against age matched controls (n = 7 each group) using BONCAT and MS | • Upregulation of TLN1, FERMT3, HIST1H4A, ILK, MPO, and VCL in FXS patients • Downregulation of AHNAK in FXS patients |
2. Mass spectrometry analysis of endogenous molecules in animal models of fragile X syndrome
2.1. Quantitative analysis of the synaptosome proteome in the Fmr1 knockout mouse model of FXS
FMRP acts as a regulator of translation for many mRNAs at the synapse, controlling the localized, activity-dependent expression of a large subset of synaptic proteins (Kim and Cho, 2014; Liao et al., 2008). In the Fmr1 knockout (KO) mouse model, the lack of negative feedback from FMRP causes dysregulation in various forms of translation-dependent synaptic plasticity. Examples include increased group 1 metabotropic glutamate receptor (mGluR) (Huber et al., 2001)- and BDNF-TrkB-mediated long-term depression (LTD) (Lessmann et al., 1994) in the hippocampus and cerebellum as well as reduced long-term potentiation (LTP) in the cortex (Dahlhaus, 2018; Kim and Cho, 2014). A key to understanding the mechanistic nature underlying synaptic dysfunction in FXS is by investigating altered synaptic protein expression in models of FXS. For this reason, many differential proteomics studies focused on analyzing the synaptosome proteome isolated from primary neuron cultures. A major benefit of analyzing sub-proteomes are that low abundant proteins are enriched, in the case of the synaptosome proteome, allowing researchers to gain a deeper understanding of the spatial and temporal processes that coordinate synaptic proteins in closely related complexes under neurotypical and disease conditions (Bai and Witzmann, 2007).
Liao, L. et al. was the first study to develop methods aimed to address synaptic protein changes in the Fmr1 KO mouse model of FXS. Here, they applied stable isotope labeling with amino acids in cell culture (SILAC)-based quantitative liquid chromatography (LC)-MS proteomics to synaptosome fractions isolated from WT and Fmr1 KO (n = 18) cortical neuron homogenates through a sucrose gradient separation method. SILAC is an approach that involves culturing cells in medium supplemented with amino acids containing light or heavy stable isotopes that are metabolically incorporated into the proteins of cells through protein synthesis (Liao et al., 2008; Zhang and Neubert, 2009). When the mixed light and heavy isotope-labeled proteins are analyzed by MS, samples can be easily distinguished due to the mass difference caused by the differential labeling; therefore, the abundance of protein is measured based on the intensities of the light and heavy peptides. The advantage to this approach is that it allows for high accuracy in quantitative data (Zhang and Neubert, 2009). In this study, 132 proteins were found to differentiate between KO and wildtype (WT) mice, including several proteins that are known to be related to autism and epilepsy, two comorbidities commonly associated with FXS (Liao et al., 2008). Many proteins found altered in the study are known or suggested to play roles in synaptic shape and cytoskeleton organization such as several members of the tissue-type plasminogen activator (t-PA) system involved in synaptic pruning; neurexin 1α, a protein involved in synapse formation and broadening; and ARVCF and lin-7, two proteins suggested to regulate synaptic maturation. This study also revealed that several proteins involved in synaptic transmission were altered in the Fmr1 KO samples. The alpha subunit of the large conductance Ca2+-activated potassium (BK) channel (Kcnma1α) was the most significantly altered (downregulated) in this group of proteins, a finding that has been reported in multiple studies of FXS (Tang et al., 2015; Zhang et al., 2014). Notably, disruptions in the Kcnma1α gene resulting in haploinsufficiency and functional deficits in BK channel activity were previously observed in a large cohort of patients diagnosed with autism spectrum disorder (ASD), suggesting a shared mechanism across different forms of intellectual disability (Laumonnier et al., 2006).
The relationship between FMRP expression and synaptic plasticity is very complex. FMRP acts pre- and postsynaptically, and its deficiently alters cellular mechanisms involved in short-term and long-term plasticity. Presynaptically, FMRP is suggested to be involved in the establishment and maintenance of synaptic connections (Christie et al., 2009). For example, the loss of FMRP in the hippocampus has been shown to affect terminal branching of mossy fibers as well as motility and dynamics of axonal growth development, altering synaptic facilitation at the presynaptic terminal (Till et al., 2011).
One group aimed to study the direct relation between the loss of FMRP and its regulated proteins in the hippocampus upon presynaptic-mediated forms of synaptic function (Klemmer et al., 2011; Till et al., 2011). Taking a quantitative proteomics approach utilizing “Isobaric Tags for Absolute and Relative Quantitation” (iTRAQ), Klemmer, P. et al. investigated changes in the hippocampus synaptic membrane proteome in Fmr1 KO mice during an early developmental period of synaptic formation and refinement. iTRAQ is a quantitative multiplexing method where each sample is labeled with a unique amine-reactive mass tag. Each isobaric tag contains three functional groups: (1) an amine-reactive group which reacts with primary amines ( NH2) located at the N-terminus of polypeptides; (2) an isotopic reporter group with distinct heavy isotope distribution (m/z 113–119 and 121); and (3) a balancer group to ensure that the mass of each tag is identical (m/z 192–186 and 184) (Zhang et al., 2016). During tandem MS (MS/MS) analysis, same-sequence peptides across all biological samples appear as a single precursor ion in MS1. Upon collision-induced dissociation peptide fragmentation, labeled peptides release their reporter ion and relative abundance across all samples is shown in MS2; therefore, protein abundance is a measure of reporter ion intensity (Zhang et al., 2016). In this study, two independent 8-plex iTRAQ experiments were carried out comparing postnatal day 14 animals from WT (n = 8) and Fmr1 KO mice (n = 8) hippocampal synaptic membranes (Klemmer et al., 2011). A total of 205 proteins with three unique peptides were quantified with 23 of these proteins being statistically significant. Proteins involved in cell differentiation and neurite outgrowth such as brain acid-soluble protein 1 (Basp1), Gap43, and cell cycle exit and neuronal differentiation protein 1 (Cend1) were upregulated in the Fmr1 KO mice (Klemmer et al., 2011). In line with these findings, the group also performed ultrastructure analysis by electron microscopy, which revealed smaller active zones with corresponding postsynaptic densities and smaller pools of vesicle clusters (Klemmer et al., 2011). A second group of proteins upregulated in the Fmr1 KO mice are involved in synaptic vesicle release such as syntaxin-1B2, synaptophysin, and synapsin-1 (Klemmer et al., 2011). In conjunction with these results, paired pulse and short-term facilitation were found to be significantly affected in the hippocampal synapses of the Fmr1 KO mice (Klemmer et al., 2011). Taken together, these findings suggested presynaptic changes underlying aberrant glutaminergic transmission in the Fmr1 KO mice during this period of development.
Another study aimed to investigate synaptic vesicle changes in the hippocampus and cerebellum of the Fmr1 KO mouse model. Synaptosome isolates from 14-week-old Fmr1 KO (n = 12) and WT (n = 11) hippocampal and cerebellar tissue were compared using label-free quantitative LC-MS and later validated using selected reaction monitoring (SRM)-MS, a targeted proteomics approach used in MS/MS where precursor ions of a particular mass are selected in MS1 and the fragmented ion products from precursor ions are selected in MS2 for detection (Broek et al., 2016). After data filtering, 23 hippocampal proteins and 13 cerebellar proteins were found to be significantly changed in the Fmr1 KO mice of which are involved in processes such as synaptic signaling, neurotransmission, synaptic vesicles, and neuron development (Broek et al., 2016). Consistent with multiple proteomic studies, this study revealed alterations in Basp1 and Sv2b (Klemmer et al., 2011; Liao et al., 2008; Xu et al., 2018). The most significantly changed proteins in both regions consisted of proteins involved in GABAergic/glutaminergic neurotransmission and ATPases; however, these proteins were downregulated in the hippocampus while being upregulated in the cerebellum (Broek et al., 2016). Notably, changes in GABAergic transmission were also correlative with previous behavior studies conducted using the Erasmus Ladder, demonstrating major deficits in associative motor learning in the Fmr1 KO mice. The group also performed live cell imaging and ultrastructure analysis, which revealed that a lack of FMRP leads to changes in synaptic vesicle unloading, with an elevated level of synaptic recycling in the cerebellum while a decrease was noted for the hippocampus (Broek et al., 2016). Elevated synaptic turn-over in the cerebellum could suggest an increase in inhibitory outputs from the cerebellum. A separate metabolomics study quantified endogenous GABA in the frontal cortex and thalamus of 5-day-old Fmr1 KO mice using LC-MS/MS in conjunction with H-magnetic resonance spectroscopy (Reyes et al., 2020). This study revealed that the lack of FMRP resulted in lower GABA concentration in thalamic and cortical regions. Taken all together, both studies suggest that aberrant GABAergic transmission is widespread and evident during multiple developmental stages in the Fmr1 KO mice(Reyes et al., 2020).
Studies have shown that FMRP regulates target mRNAs in a cell type-specific and developmentally regulated manner (Li et al., 2020), which is also reflected when investigating changes at the proteome level. One synaptosome proteomics study aimed to identify age-dependent changes in protein expression by comparing two developmental time points in the Fmr1 KO mouse against the same number of age-matched WT mice: postnatal day 17 (n = 4), a period of critical synaptogenesis, and adulthood (postnatal day 45; n = 4) (Tang et al., 2015). The researchers used stable isotope labeling in mammals (SILAM) and high-resolution MS to quantify proteins from neocortical synaptosome-enriched samples. SILAM is a similar method to SILAC except SILAM involves feeding the mice a N-enriched diet, allowing for complete labeling of the whole animal, whereas SILAC involves supplementing cell culture medium with heavy- and light-labeled amino acids. This study resulted in the identification of 2600 proteins of which 999 proteins (P < 0.05) were found to be differential between WT P17 mice and Fmr1 KO P17 mice. The number of differential proteins between WT and Fmr1 KO in P45 mice was significantly lower suggesting that proteins involved in FXS pathogenesis are present at a young age during periods of synaptic refinement. In total, 15 proteins were selected for further validation by Western blotting, which comprised of core proteins in presynaptic vesicles (synapsin 1 and 2, SNAP 25); postsynaptic channels and receptors (Hcn1, mGlu7); signaling molecules (CaMKIIa, Darpp32); synaptic scaffolding proteins (Shank family proteins-Shank 1, Shank2, Shank3, PSD95, Dpsyl3); adhesion molecules (Chl1) and metabolic enzymes (Hsd11b1). Apart from Chl1, Dpsyl3, and mGluR7, many of these proteins were significantly upregulated in the P17 Fmr1 KO mice. The group determined that the elevation of proteins is correlated to FMRP deficiency though a separate LC-MS/MS study on WT and Fmr1 KO neurons where they stimulated translation by treating neurons with a mGluR agonist, (S)-3,5-dihydroxyphenylglycine (DHPG) (Tang et al., 2015). The results showed that neurons derived from the Fmr1 KO mice produced nascent synaptic proteins at a higher rate than WT neurons, in which many of these proteins were previously found to be direct mRNA targets of FMRP (Tang et al., 2015) (Darnell et al., 2011).
2.2. Homeostatic plasticity in the Fmr1 KO mouse model: Steady state and activity-dependent proteome changes
Homeostatic plasticity is crucial for the refinement and maintenance of functional neural networks and represents a series of molecular cascades that sense, respond, and drive changes in response to changes in physiological activity levels. While disruptions in protein synthesis have been documented in studies of FXS (Jacquemont et al., 2018), a question that remains unsolved is whether these disruptions occur during homeostatic conditions or is this the result of inappropriate responses to activity. For this reason, a few studies have aimed to address this concern by analyzing proteome changes under steady state and stimulated environments.
Bowling, H. et al. performed a comprehensive study on acutely prepared dorsal hippocampal slices from 10 to 12-week-old Fmr1 KO mice and their WT littermates (n = 9 WT, n = 9 KO pooled into 3 groups of 3 mice per pool) to compare steady state differences in translation between KO and WT groups as well as in response to stimulation of mGluR5 signaling using DHPG (Bowling et al., 2019). To measure de novo protein synthesis, the group performed BONLAC proteomics three separate times for a total of three independent biological replicates per group in conjunction with SILAC labeling to measure protein abundance and control for any possible labeling bias. BONLAC proteomics is a quantitative de novo protein synthesis-labeling technique that uses click chemistry biorthogonal noncanonical amino acid tagging (BONCAT) to identify de novo peptides (Bowling et al., 2016). After data analysis, the study revealed 324 altered newly synthesized proteins at steady state noting the presence of both up- and down-regulated proteins; however, up-regulated proteins showed a higher-fold change (Bowling et al., 2019). Gene ontology analysis indicated that the top enriched functional clusters in the dataset were biosynthesis, protein-folding, synaptic proteins, and neurodegeneration-related proteins (Bowling et al., 2019). Specifically, the group validated three high confidence candidate proteins differently expressed in the Fmr1 KO group using BONCAT-specific pull-downs: mGluR5, upregulated; NMDAR2B/Grin2B, downregulated; and SynGAP, upregulated (Bowling et al., 2019). Analysis of the DHPG-stimulated results revealed 278 differential proteins of which 164 were upregulated and 86 were downregulated. Interestingly, the data showed that the de novo proteomes under steady state and stimulated condition had limited overlap, showing that major categories of the differential proteins under stimulated conditions were cell membrane and biosynthesis proteins (Bowling et al., 2019). The group validated two proteins, Ras and hexokinase 1, in which both were downregulated in the Fmr1 KO mice, a finding that disagrees with Licznerski, P. et al., which noted an elevated level of hexokinase 1 in FX neurons; however, this inverse in expression may be due to DHPG stimulation conducted in Bowling, H. et al. Findings from this study suggests that the disruptions in protein synthesis in the FXS mouse model occurs both at steady state and in response to mGluR5 stimulation.
Alternatively, a different group aimed to understand the steady state proteome changes after chronic activity deprivation achieved by treatment with tetrodotoxin (TTX) and an NMDA receptor antagonist, (2R)-amino-5-phosphonovaleric acid (APV), in WT and Fmr1 KO cortical neurons dissected from P0-P1 mice (Bülow et al., 2021). TTX-APV treatment was selected for this study since their previous study revealed that this drug treatment induced intrinsic homeostatic plasticity in Fmr1 KO neurons (Bülow et al., 2019). Using Tandem Mass Tagging (TMT) quantitative mass spectrometry, the group identified 6074 proteins using a 22-plex experimental design to simultaneously compare vehicle or TTX-APV treated WT and Fmr1 KO neurons (Bülow et al., 2021). TMT quantitative MS applies similar chemistry like iTRAQ in that they are both multiplexing techniques that utilize isobaric tags; however, TMT isobaric tags are composed of different functional groups. After data analysis, the study revealed that drug treatment induced changes in both WT and Fmr1 KO neuron cells; however, Fmr1 KO cells were more sensitive to TTX-APV treatment with 159 proteins showing expression change in WT cells and 260 proteins showing changes in Fmr1 KO cells. Of the proteins sensitive to activity blockade, many localized to the mitochondrial membrane and play roles in the tricarboxylic acid cycle and respiratory electron transport including Ndufa11, Mpc2, Atp5d, and Cox14 (Bülow et al., 2021). Overall, the main conclusions drawn from the study are (1) the mitochondrial proteome was most affected during chronic activity deprivation, and (2) Fmr1 KO neurons show exaggerated changes in the mitochondrial proteome during activity deprivation (Bülow et al., 2021). Together, these findings suggest that FMRP attenuates activity dependent modification of the mitochondrial proteome.
Another group interested in investigating activity dependent changes in NMDARs performed quantitative MS and in vitro stimulation of isolated mouse synaptoneurosomal to investigate local protein synthesis in 1–2-month-old Fmr1 KO against age-mated WT mice (n = 4 per group) (Kuzniewska et al., 2020). To promote induction of NMDAR, mouse synapses were treated for 30 s with NMDA and glutamate with the addition of a selective NMDAR antagonist (APV) to avoid excitotoxity on the synapses (Kuzniewska et al., 2020). The same biological samples were measured using two different quantitative methods, iTRAQ8 and TMT10, and in total 2080 proteins were quantifiable with 516 proteins being significantly upregulated in the stimulated synaptoneurosomal samples (Kuzniewska et al., 2020). Similarly, to what was observed in the previously mentioned study on NMDAR blockade, the mitochondrial proteome was heavily affected after NMDAR stimulation. Specifically, 393 of all 2080 quantified proteins and 69 of the 516 upregulated proteins locally synthesized at the synapse were mitochondrial proteins that are incorporated into respiratory chain complexes (Kuzniewska et al., 2020). The group also performed high-resolution respirometry using multiple respiratory substrates to target specific respiratory chain complexes. Fmr1 KO mitochondria show elevated levels of respiration, determined by oxygen consumption rates, in the presence of succinate (complexes II-IV) and ascorbate and TMPD (complex 4) (Kuzniewska et al., 2020).
How the loss of FMRP triggers changes in the mitochondrial proteome is not fully understood; but, given that only 18 mitochondrial proteins are mRNA bound and translationally regulated by FMRP determined by comparing a list of mRNA targets of FMRP against the MitoCarta dataset (Darnell et al., 2011; Rath et al., 2021), one can suggest that FMRP influences the mitochondrial proteome through an indirect FMRP-dependent mechanism. Licznerski, P. et al. conducted MS proteomics and directly investigated cellular metabolic changes in neurons derived from Fmr1 KO mice. The researchers discovered that these neurons exhibit a mitochondrial inner membrane leak, leading to abnormal synaptic development (Licznerski et al., 2020). To understand the implications of this leak metabolism, the researchers also examined human FXS fibroblasts. They found that closing the leak channel of the ATP synthase, a key enzyme in mitochondrial energy production, normalized the translation rate of mRNA, reduced lactate levels, and restored the activity of glycolytic and tricarboxylic acid (TCA) cycle enzymes, which were found to be dysregulated based on their proteomics data (Licznerski et al., 2020). Interestingly, other proteomic studies in the Fmr1 KO mouse model conducted under different experimental conditions also observed direct changes or factors that could be influencing mitochondrial dynamics in the Fmr1 KO mice. One study applied MALDI-MS imaging and LC-MS/MS to study the effects of a soy protein-based diet in the Fmr1 KO mice (Ma et al., 2024). This study observed that differential proteins expressed in the hippocampus and hypothalamus of Fmr1 KO mice on a soy protein-based diet versus the mice on a casein protein-based diet are significantly associated with mitochondrial metabolism such as glycolytic and tricarboxylic acid enzymes (Ma et al., 2024), suggesting possible dietary influences on mitochondrial metabolism. A study conducted by Talvio, K. et al. applied inductively coupled plasma-MS (ICP-MS) to study trace elements in Fmr1 KO mice, which revealed increased iron contents in the heart and cerebellum. Iron, a redox-active metal, is critical for many proteins involved in mechanisms that defend against oxidative stress (el Bekay et al., 2007); however, accumulation of iron can hinder the ability of neuronal cells to respond to oxidative stress as this has been linked to pathophysiological events such as abnormal cerebellar myelination (Talvio et al., 2021). With studies in the Fmr1 KO mice showing a deficient antioxidant system and increased levels of reactive oxygen species (el Bekay et al., 2007), this can contribute to the alterations seen in the mitochondrial proteome.
Another possible influencing factor could be related to abnormalities in NMDARs. Exaggerated mGluR5 signaling is a well-supported observation in FXS, and consequently, this was shown to induce NMDAR dysfunction in Fmr1 KO mice (Aloisi et al., 2017). The relationship between mGluR5 and NMDAR has been studied many times, and it is now understood that mGluR5 potentiates ionotropic NMDAR activity by regulating trafficking and subcellular distribution of NMDARs, GluN1 and GluN2B, through pathways involving CaMKII (Jin et al., 2015). Also, mGluR5 and NMDAR co-assemble in the same post-synaptic density complex. Specifically, the presence of Homer1a permits direct interactions between NMDAR and mGluR5, which facilitates mGluR5-mediated inhibition of NMDAR currents (Aloisi et al., 2017). In FXS, the interaction between the two receptors was observed to be dysregulated. The group investigating this highlighted three key observations: 1) Impaired scaffolding of mGluR5/Homer alters mGluR5 diffusion along the membrane; 2) Fmr1 KO neurons exhibit enhanced mGluR5/NMDAR co-clustering; and 3) tighter mGluR5/NMDAR clustering reduces NMDAR plasticity (Aloisi et al., 2017). How their interactions tie into mitochondrial dysfunction has yet to be explored in FXS; however, a study published using PC12 cells, derived from adrenal pheochromocytoma, found that mGluR5 activation attenuates NMDA-induced neurotoxicity and mitochondrial dysfunction by disrupting the interaction between GluN2B/NMDAR and PSD95 (Dai et al., 2014). This was observed to prevent the generation of reactive oxygen species and the release of cytochrome C from the mitochondria (Dai et al., 2014). Given that pharmacological manipulation of NMDARs led to alternations in the mitochondrial proteome in the Fmr1 KO mice neurons, these changes could result because of aberrant mGluR5/NMDAR signaling.
2.3. Analysis of post translational modifications: Site specific modifications on FMRP and global changes in Fmr1 KO mice
Post translational modifications (PTMs) of proteins are molecular regulatory mechanisms that drive many biochemical and cellular processes and studying the diversity of PTMs is crucial for understandings mechanisms that influence cellular regulation. MS is a fundamental tool for the detection and quantitation of these covalent modifications (Witze et al., 2007). Modern approaches in MS make it possible to conduct high throughput experiments that allow researchers to globally screen for changes in various PTMs, and in turn, gain a deeper understanding to how protein chemistries impact biological functions (Witze et al., 2007).
Early research efforts on FXS focused heavily on understanding the function of FMRP and how this protein is regulated. The pioneering work of Ceman, S. et al. gave us great insight to the phosphorylation dynamics of FMRP. Here they applied MALDI-time of flight (TOF) MS and site-directed mutagenesis to determine phosphorylation sites on FMRP purified from mouse fibroblast L-M(TK-) cells and mouse brain lysates. First, L-M(TK-) cells were metabolically labeled with [32P] and isolated via Flag-tagged FMRP by immunoprecipitation with an anti-Flag antibody to confirm whether mammalian FMRP is a phosphoprotein (Ceman et al., 2003). Precursor ion scanning of the tryptic digest FMRP from the transfected L-M(TK-) cell line revealed phosphorylation in a region of FMRP between amino acid residues 483–521 with one to three phosphoresidues (Ceman et al., 2003). After narrowing down the region for phosphorylation, MS/MS sequencing analysis was carried out on purified FMRP from mouse brain lysates to reveal four possible kinase substrates: S496, T501, S503, and the high conserved S499 residue (Ceman et al., 2003). Applying site-directed mutagenesis to replace serine 499 with alanine or aspartic acid, the group observed that aspartic acid, but not alanine, was able to partially restore phosphorylation even though aspartic acid is not a kinase substrate (Ceman et al., 2003). This suggested that a negative charge at position 499 triggers phosphorylation of a second site between residues 496–503 in a hierarchical manner. This study also revealed that the phosphorylation status of FMRP on S499 had no effect the association with RNAs or its translocation; however, it did influence the translation state of FMRP-associated polyribosomes by showing that phosphorylated FMRP associated with stalled polyribosomes while unphosphorylated FMRP associated with actively translating polyribosomes (Ceman et al., 2003). The work published by Ceman, S. et al., which was later replicated by Bartley, C. et al. (Bartley et al., 2016) confirming these initial findings, provided a framework for identifying kinases and phosphatases that regulate the phosphorylation status of FMRP.
A few other proteomics studies have employed more modern MS protocols to study global PTMs in the Fmr1 KO mouse model of FXS. Matic, K. et al. performed quantitative analysis on the proteome and phosphoproteome on Fmr1 KO mouse embryonic fibroblasts (MEFs) using high resolution MS and SILAC labeling. When studying PTMs, typically samples are processed in a way that enriches proteins or peptides that contain the desired PTM. The technique employed in this study was metal oxide affinity chromatography (MOAC). In MOAC, negatively charged phosphate groups on the phosphopeptide are selectively bound to metal oxide. TiO2 is commonly used as it shows strong binding affinity for phosphorylated peptides (Qiu et al., 2020). The proteome analysis revealed 4195 quantified proteins with 266 proteins showing significant changes (134 increased; 132 decreased). Analysis of the phosphoproteome revealed 6040 phosphorylation events on 2494 proteins after normalizing phosphopeptide ratios with protein ratios, and of these phosphorylation events, 142 showed significant changes with 86 increasing and 56 decreasing phosphorylation sites in the Fmr1 KO cells (Matic et al., 2014). Two high confidence proteins from the proteomics data were validated by western blot noting a significant decrease in the major prion protein (Prp), a protein involved in synapse formation, and a significant increase in cytosolic-dependent phospholipase A2 (cPLA2), a protein that participates in cerebellar LTD and motor learning (Matic et al., 2014). The group analyzed the 266 significant proteins and 142 phosphorylation events in the context of major signal transduction pathways. In the Fmr1 KO cells, the MEK/ERK pathway was downregulated with decreased phosphorylation of ERK1/2, IRIS, and Rictor and decreased protein levels of PTEN, the PI3K-Akt inhibitor (Matic et al., 2014). Other differential proteins significant in the Fmr1 KO cells include proteins involved in Wnt signaling (e.g., Knypek, decreased; PLC, decreased; PKC, decreased phosphorylation), p53 signaling (e.g., Chk1, Cdk4/6, cyclin B, Cdc2, all increased; PTEN, TSAP6, both decreased), and MAPK signaling (e.g., EGF-receptor, Ras, and ERK1/2, all decreased) (Matic et al., 2014). Together, these findings suggest disruptions in phosphorylation events in Fmr1 KO MEF cells and warrants further investigation in other cell types such as neuronal cultures.
MS approaches are not limited to enrichment of phosphopeptide. Strategies can be applied to studying other PTMs such as glycosylation, acetylation, methylation, etc. Lysine acetylation (K-ac) is a reversible, global PTM commonly associated as a PTM on histones that regulates gene transcription through chromatin remodeling. In recent years, evidence suggests that K-ac in non-histone proteins plays a role in many cellular functions such as regulation of metabolic pathways, enzyme activity, and subcellular localization of proteins (Narita et al., 2019). A recently published study aimed to investigate the lysine acetylome in the hippocampus of Fmr1 KO mice. Using TMT quantitative mass spectrometry to analyze peptides enriched by anti-K-ac antibodies, this study identified a total of 1629 K-ac peptides on 717 proteins with a 6-plex experimental design (n = 3 for Fmr1 KO and WT groups). In total, 51 K-ac peptides localized on 45 proteins were significantly changed (25 downregulated, 26 upregulated) in the Fmr1 KO mice most of which were associated with metabolic control and synaptic function (Wu et al., 2022). Enrichment analysis of these significant K-ac proteins identified nine enriched functional categories with the top three being carboxylic acid metabolism, aerobic respiration, and the tricarboxylic acid cycle (Wu et al., 2022). Four proteins common among all three functional categories were dihydrolipoamide dehydrogenase (DLD, upregulated), Pyruvate dehydrogenase E1 component subunit alpha (PDHA1, downregulated), Succinate dehydrogenase (SDHA, downregulated) and Succinate-CoA ligase alpha subunit (SUCLG1, upregulated) (Wu et al., 2022). Since much of the data pointed to dysregulated energy metabolism, the researchers assessed levels of ATP and lactate in the mouse hippocampus and cultured HT22 hippocampal cells. The results showed that ATP levels in the Fmr1 KO hippocampi and KO cells were significantly lower than the control group, and the inverse was shown when measuring lactate with the Fmr1 KO groups showing significant increase than the controls. The data presented in the study suggest that the deficiency of FMRP and alterations in K-ac proteins could contribute to abnormal mitochondrial function and metabolism.
2.4. Analysis of differential proteins in the Drosophila model of FXS
The Drosophila model of FXS (dfmr1 null mutants) was first established in 2001 (Zhang et al., 2001) to complement research conducted in the Fmr1 KO mouse model. Even though this model lacks the complexity of the mouse model, Drosophila FMRP (dFMRP) contains the defined functional domains and PTMs as human/mouse FMRP and exhibits similar subcellular expression patterns and regulatory roles as observed in the mouse models; however, the Drosophila genome contains a single conserved fragile-X related (dfxr) gene corresponding to mouse and human FMR1, FXR1, and FXR2, suggesting dfxr mediates the conserved functions of all three genes (Zhang et al., 2001). A few proteomics studied have been conducted in dfmr1 null mutant models, and while the proteome has been less explored in the Drosophila model in comparison to the Fmr1 KO mouse model, these studies offer a unique evolutionally perspective to the conserved functional roles of FMRP.
Zhang, Y. et al. analyzed the brain proteome of 75 2-day-old dfmr1 null mutants against 75 WT age-matched controls conducted as three independent experiments (25 dfmr1 null mutants; 25 WT controls) using two-dimensional difference gel electrophoresis (DIGE) and MALDI-TOF MS for protein identification. In total, 1500 proteins were identified with 24 proteins showing significantly altered expression levels of a 1.3-fold or greater increases or decreases (Zhang et al., 2005). The differentially expressed proteins were noted to be involved in processes such as energy metabolism, monoamine synthesis, protein degradation, redox and iron homeostasis, and cytoskeleton organization (Zhang et al., 2005). Interestingly, the researchers suggested many protein expression changes in the dfmr1 mutant brain may not be directly due to dFMRP meditated translation but rather indirect interactions due to altered PTMs. For example, four different isoforms of glycerol-3-phosphate dehydrogenases were found altered with two isoforms showing major increases and two showing major decreases (Zhang et al., 2005). This observation was also seen in the two proteins involved in monoamine synthesis, phenylalanine hydroxylase/Henna and GTP cyclohydrolase 1/Punch. Both proteins had multiple isoforms with at least one isoform showing an increase and another one showing a decrease in expression (Zhang et al., 2005). Phenylalanine hydroxylases, encoded by the Drosophila henna gene, and GTP cyclohydrolase, encoded by the Drosophila punch gene, are in the same pathway for the biogenic amine synthesis of dopamine and serotonin, in which levels for each were quantified, showing dfmr1 null mutants have 80% elevated levels of dopamine and 30% elevation in serotonin. The data also revealed a series of cytoskeleton proteins such as β-tubulin and actin 5C, both of which were elevated in the dfmr1 null mutants. Previous proteomic studies conducted by the group showed that the loss of dFMRP alters microtubule cytoskeleton stability during spermatogenesis in the testes (Zhang et al., 2004) and synaptogenesis in the brain through regulating the translation of the Drosophila homolog of MAP1B, Futsch. In the dfmr1 mutants, Futsch was noted to have a 2-fold increase in expression levels (Zhang et al., 2001). Based on the findings from their previous work and this current study, the results can predict that the microtubule cytoskeleton network is aberrantly hyperstabilized in the dfmr1 mutants.
Alterations in cytoskeletal proteins were also seen to have to effects on cleavage furrow formation in dfmr1 mutants, a form of cytokinesis where cortically positioned nuclei are encapsulated by plasma membrane furrows. This process was revealed to be highly dependent on reorganization of actin and microtubule cytoskeletons in a study conducted by Monzo et al. on dfmr1 mutant embryos. Using 2D DIGE/MALDI-TOF MS, this study revealed that three subunits (CG5525, Ccty, and Tcp-Iη) of the chaperonin containing TCP-1 (CTT) complex were misregulated in dfmr1 mutants (Monzo et al., 2010). This study suggests that dfmrp is required for the coordinated expression of various subunits within a multi-subunit complex. The use of DIGE in the dfmr1 null mutant studies presents an evident limitation since the proteins revealed through this technique are limited to proteins with isoelectric points between pH 4–7 and molecular weights between approximately 15–150 kDa. This warrants further investigation using newer proteomics approaches that allow for greater resolution of the dfmr1 null mutant proteome.
2.5. Analysis of FMRP-targeted mRNAs and differential proteins in Fmr1 KO mice
Proteomic studies on models of FXS offer avenues to answer key questions to the molecular biology of FXS. One of the more pressing questions is whether FMRP-targeted mRNAs are dysregulated at the protein level, and if so, do they present with similar cell-type specificity characteristics as FMRP-targeted mRNAs. In efforts to investigate these questions, we performed a comparative analysis on FMRP-targeted mRNAs and significantly differential proteins from the studies mentioned in this review. Supplementary files from CLIP studies that aimed to identify translationally regulated FMRP targets (Darnell et al., 2011; Hale et al., 2021; Maurin et al., 2018; Sawicka et al., 2019) were compared against significantly altered proteins from MS studies on Fmr1 KO mice brain tissue (Bowling et al., 2019; Broek et al., 2016; Bülow et al., 2019; Klemmer et al., 2011; Kuzniewska et al., 2020; Liao et al., 2008; Seo et al., 2022; Tang et al., 2015; Xu et al., 2018). To investigate whether any differential proteins exhibit distinct patterns in different cell types, we divided these studies into cortical or hippocampal groups based on the brain region analyzed in Fmr1 KO mice.
Across the five studies that conducted proteomics on cortical lysates in Fmr1 KO mice (Broek et al., 2016; Bülow et al., 2019; Kuzniewska et al., 2020; Liao et al., 2008; Tang et al., 2015), 217 significantly upregulated proteins and 58 significantly downregulated proteins were found to be FMRP targets. GO analysis of the 217 upregulated proteins showed top enrichment for processes such as vesicle-mediated transport in synapse (e.g., Snap25, Ap2a2/b1/a1, Ctnnb1), synaptic vesicle cycle (e.g., Camk2a, Bsn, Bin1, Syn1), synapse organization (e.g., Homer1, AhNK3, App, Tuba1a) and modulation of chemical synaptic transmission (e.g., Grin2a, Map1b, Mapk1), whereas downregulated cortical proteins showed enrichment for dendritic spine development (e.g., Cask and Mtor) postsynapse organization (e.g., Snx27, Actb) and RNA splicing (e.g., Hnrnpa2b1 and Dhx9) (Fig. 1). There were 26 additional cortical proteins that were not included in the enrichment analysis since they were found to be up-and down-regulated across multiple studies. For example, in two separate studies from the same group, Shank2, Sv2a, and Syt1 were all downregulated in the Liao et al. study, but were all significantly upregulated in the study by Tang et al. We speculate the difference in synaptic protein expression may be due distinct developmental timepoints since Liao et al., conducted proteomics on embryonic day 18 cortical neuron synapses, a critical timepoint for neurogenesis, whereas in Tang et al., neocortical synaptic fractions were from P17 mice, a period of peak synaptogenesis in mice. Interestingly, pathway analysis of the upregulated cortical proteins enriched cGMP and cAMP signaling, which was one of the most prominent deregulated pathways in the HITS-CLIP study of Maurin et al.; however, in their study, they postulated that the elevation of PDE2A in the cortex and hippocampus would imply a reduced level of cAMP and cGMP (Maurin et al., 2018). Additionally, Wnt signaling pathway was enriched in the analysis of down regulated cortical proteins, which has been previously reported to be reduced in neural progenitor cells due to FMRP loss and affect neurogenesis (Luo et al., 2010).
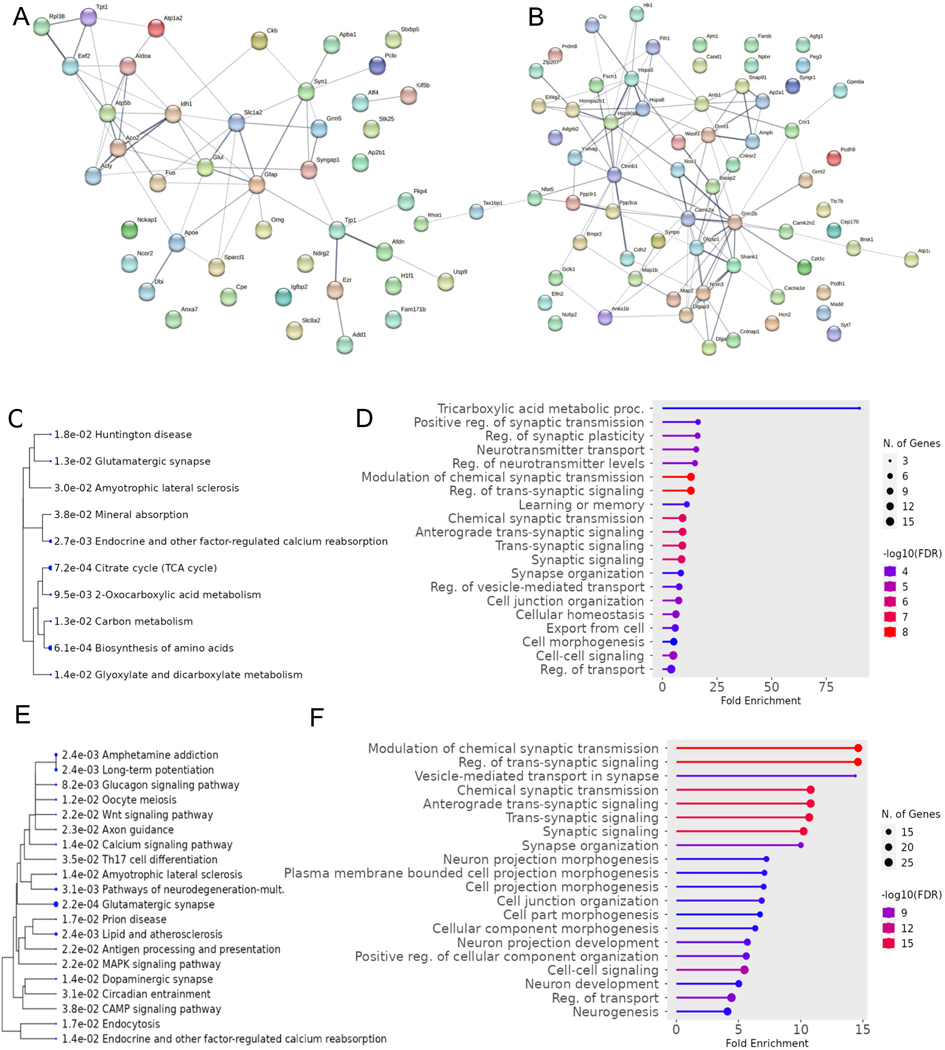
Analysis of proteomic studies on hippocampal samples of Fmr1 KO mice show partial overlap with cortical proteins that transcriptionally regulated by FMRP. There were 23 proteins that were upregulated in bother regions (e.g., Glul, Syngap1, Fus, Slc8a2) and Map2, Madd, Hnrnpa2b1, and Hspa5 were the only four shared downregulated proteins. Notably, a significant portion of the downregulated hippocampal proteins were found to upregulated in the cortex such as Grin2b, Camk2a, Shank1, and Synpo; however, it is unclear whether this divergence is due to the difference in experimental conditions between studies or related to FMRP loss. In total, 47 upregulated proteins and 68 downregulated proteins expressed in the hippocampus across four proteomic studies (Bowling et al., 2019; Broek et al., 2016; Klemmer et al., 2011; Seo et al., 2022) were matched to datasets on FMRP targets. Upregulated proteins were enriched for processes such as tricarboxylic acid metabolic processes (e.g., Idh1, Acyl, Aco2) regulation of synaptic transmission and plasticity (e.g., Glul, Gfap, Kif5b, Dbi), and neurotransmitter transport (e.g., Pclo, Stxbp5, Apba1), whereas downregulated proteins were enriched for processes such as vesicle-mediated transport (e.g., Dmn1, Syt7, Nrxn3) trans-synaptic signaling (e.g., Dlgap1, Ppp3r1/ca, Nos1, Synpo), and neuron projection morphogenesis (e.g., Cdh2, Map2/1b, Shank1) (Fig. 2). Interestingly, in Sawicka et al., FMRP was found to regulate circadian transcripts in CA1 neurons. The pathway analysis of proteins downregulated in hippocampus revealed enrichment for MAPK signaling pathway and circadian entrainment. MAPK pathways can function as inputs allowing the endogenous clock to entrain to 24 h environmental cycles (Goldsmith and Bell-Pedersen, 2013). Furthermore, nitric oxide synthase 1, a FMRP regulated transcript that functions to synthesize nitric oxide from L-arginine, was notably downregulated in the hippocampal proteome of Fmr1 KO mice in Seo et al. Since nitric oxide plays an important role in phase-shifting of circadian neuronal activities (Ko et al., 2013), this may provide insight to mechanisms underlying sleep disorders in individuals with FXS (Carotenuto et al., 2019).
Fig. 2. Bioinformatic Analysis of Differential Hippocampal Proteins Translationally Regulated by FMRP.
A) Protein-protein interaction network of upregulated hippocampal proteins. B) Protein-protein interaction network for downregulated hippocampal proteins. C) Pathway enrichment for upregulated hippocampal proteins. D) GO enrichment for upregulated hippocampal proteins. E) Pathway enrichment for downregulated hippocampal proteins. F) GO enrichment for downregulated hippocampal proteins. Analysis was done in ShinyGo 0.80 and String (Ge et al., 2020).
3. Mass spectrometry proteomics in human models and human tissue samples of fragile X syndrome
Although many promising protein targets have been identified in mouse model studies, the genetics of the Fmr1 KO mice lacks a fundamental aspect that occurs in humans. The Fmr1 mutation in KO mice does not recapitulate the timing of developmentally regulated embryonic events that leads to transcriptional silencing of FMR1 since mice do not have a functional Fmr1 gene, whereas in humans, FMRP is present during early gestation and then becomes substantially reduced or absent after hypermethylation when maternal epigenetic signatures are enacted. With this difference at play, validation of data from Fmr1 KO mice in human samples of FXS is a critical need for the FXS field in order to make progress towards identifying protein targets for therapeutic intervention. Only a limited number of proteomic studies have been conducted in human models of FXS, mainly due to limitations regarding sample collection and accessibility. These studies, for the most part, can be placed into four categories: development of diagnostic tools, proteinprotein interaction studies, multiomic approaches, and human tissue analysis and will be the focus for the remainder of this review.
3.1. Mass spectrometry as a diagnostic tool for fragile X syndrome
Methods for diagnosing FXS are standardized and typically involve analyzing DNA and protein isolated from patient blood or buccal samples using three main techniques: FMR1 PCR-capillary electrophoresis to determine CGG repeat length, Southern blot for repeat length and methylation profiles, and a Luminex-based immunoassay for FMRP quantification. A few limitations regarding current standard laboratory testing for FXS are that PCR assays only target repeat size and do not provide information on methylation state of the FMR1 promoter, necessitating further testing using methylation sensitive Southern blot; however, this method is relatively low throughput. Recently sensitive methylation PCR methods have been developed, particularly an assay developed at Asuragen (Chen et al., 2011) and are utilized in some clinical laboratories instead of Southern blot. Though not proteomic-based, Godler, D. et al. presented a novel approach using MALDI-TOF MS for FMR1 locus methylation mapping to overcome these issues. For this study, blood samples from controls (n = 49), gray zone carriers (40–54 CGG repeats; n = 18), premutation carriers (55–170 CGG repeats; n = 22), and FXS subjects (n = 22) were processed for DNA extraction, and the remainder of the protocol was based on in vitro transcription of bisulphate-converted DNA, CGG repeat size PCR amplification, base-specific cleavage, and fragmentation analysis using MALDI-TOF MS (Godler et al., 2010). Using this methodology, the group was able to map methylation patterns of five target regions (amplicons 1–5) consisting of the FMR1 promoter, as well as the 5′ and 3′ adjacent regions. Amplicon 2 encompasses the main FMR1 transcriptional start site next to the CGG expansion. Two novel regions, amplicon 1 and 5 referred to as fragile X-related epigenetic element 1 and 2 (FREE1 and FREE2, respectively), were chosen for further investigation since these regions appeared to hypermethylated in FXS individuals but unmethylated in carriers with smaller expansions (Godler et al., 2010). The potential regulatory rolls of FREE1 and FREE2 regions are supported by two observations: 1) a positive correlation between methylation status of FREE1/FREE2 and amplicon 2 in high functioning males, including FXS individuals with mosaicism, and 2) negative correlations between FREE1/FREE2 and the number of FMRP-positive lymphocytes and the FMR1 activation ratio in blood of high functioning males and females with variable FMR1 activation(Godler et al., 2010).
A series of studies were published utilizing this methodology to investigate the clinical application of FREE1 and FREE2 methylation analyses. For females, Southern blot analysis provides the activation ratio, which is the proportion of unmethylated alleles on the active X chromosome. One study found that FREE2 methylation analysis was a better predictor than activation ratio to determine the proportion FMRP positive cells in blood in 22 females with FXS (Godler et al., 2011). In line with these findings, the group investigated whether methylation patterns on certain CpG sites within the FREE2 region could be a predictor of cognitive status in females with FMR1 mutations. For this study, blood samples from 72 controls, 62 premutation females, and 18 full mutation females with previously conducted Wechsler intelligence quotient (IQ) tests were analyzed using MALDI-TOF MS and methylation patterns of the FREE2 region was analyzed. The results showed that methylation of intron 1 CpG sites 10–12 showed the highest diagnostic sensitivity and specificity (100% and 98%, respectively) for detecting females with a standardized verbal IQ of <70 (Godler et al., 2012). In the full mutation cohort, methylation of these sites correlated significantly with full-scale, verbal, and performance IQ (Godler et al., 2012).
Zhang, J. et al. conducted a study aimed to characterize the proteoforms of FMRP. For this study, FMR1 cDNA clone was transfected into human HEK293 cells to express full length human FMRP. Purified FMRP was digested and analyzed by LC-MS/MS. The group was able to characterize two additional FMRP protoforms, isoform 4 and isoform 7, in addition to the full length FMRP isoform, isoform 6 (Zhang et al., 2019). FMRP isoform 4 and isoform 7 share the same N-terminal sequence with isoform 6; however, isoform 4 and isoform 7 have C-terminal truncations of 25 and 17 amino acid residues relative to isoform 6, respectively (Zhang et al., 2019). This also corresponds to the differences in molecular weight between the isoforms with isoform 4 having a molecular weight of 68 kDa and isoform 7 having a molecular weight of 69 kDa in comparison to full length 73 kDa FMRP (Zhang et al., 2019). Data analysis revealed high sequence coverage for isoform 4, 6, and 7 suggesting these isoforms are the most abundant FMRP splicing isoforms in human HEK239 cells (Zhang et al., 2019). The focus of this work was devoted to quantification of tryptic peptides of FMRP, which could provide an important basis to the development of targeted FMRP quantification by MS for measuring FMRP in human blood samples. The study revealed that peptides SFLEFAEDVIQVPR and MEELVVEVR provided the highest spectral counts and represent the most sensitive peptides for targeted quantification analysis of FMRP in clinical samples (Zhang et al., 2019). Advantages to MS-based targeted assays over conventional Luminex assays include higher analytic throughput, high specificity for proteins with similar sequence variants, and the potential to multiplex. The data provided by the authors support that MS-based assays are clinically relevant, high throughput tests, but further validation in larger independent cohorts need to be conducted before being implemented in clinical routine.
3.2. Analysis of protein-protein interaction networks: The FMRP interactome
The most prominent and studied function of FMRP is its role in translation regulation. Alongside that, FMRP plays critical roles in mRNA transport, chromatin dynamics, and RNA binding. However, the underlying molecular mechanisms and networks that regulate and guide FMRP remains elusive. For this reason, a few proteomic studies aimed to identify subcellular FMRP protein networks, which would allow researchers to gain insight into the roles of FMRP at the post-translational layer of regulation and thus has potential to reveal novel functions of FMRP.
Taha, M. et al. conducted affinity pulldown experiments using HeLa cell lysates and purified GST fusion protein fragments of the N-terminal, central, and C-terminal domains of human FMRP. Proteins bound to N and C termini domains were used for analysis since these are the main locations for FMRP protein-protein interactions. Pull-down samples were separated via SDS gels and each fraction, GST-FMRPN-term, GST-FMRPC-term, and GST-control, was process for LC-MS/MS analysis. The experiment revealed a total of 102 FMRP interacting proteins with 22 localized to the N-terminal domain, 67 localized to the C-terminal domain, and 13 proteins associated with both N- and C-terminal domains (Taha et al., 2021). Categorially, most FMRP-interacting proteins are involved transcription (e.g., transcription intermediary factor 1β, TRIM28; Myb-binding protein 1 A, MYBBP1A), RNA metabolism (e.g., nuclear protein homolog 2, NOP2; DEAD box protein 5, DDX5), and translation (e.g., elongation factor 2, eEF2; 60s ribosomal protein L36, RPL36). Two proteins, non-POU domain-containing octamer binding protein (NONO) and RAS GTPase-activating protein (G3BP1) were two proteins chosen for further analysis. Both proteins were previously described at components of RNP granules, specifically stress granules, which are aggregates of untranslated mRNAs sorted between decay, storage, or polysome assembly (Kedersha and Anderson, 2009). Using structured illumination super-resolution microscopy, the group was able to show that FMRP and G3BP1 colocalize in stress granules (Taha et al., 2021). A study mentioned above by Zhang, J. et al. also revealed several FMRP interacting proteins in HEK293 cells though the focus of this study was to characterize different proteoforms of FMRP. A few proteins showed consistencies with the FMRP interacting proteins in HeLa cells published by Taha, M. et al. such as Bcl-2-associated transcription factor 1 (BCLAF1), nuclear FMR1-interacting protein 2 (NUFIP2), which was initially discovered in 2003 as a FMRP interacting protein (Bardoni et al., 2003), and RNA binding protein FXR1 (Zhang et al., 2019), the autosomal homolog of FMRP first described in 1995 (Siomi et al., 1995).
While the interactome studies of Taha et al., and Zhang et al., give us insight to FMRP-interacting protein networks, we cannot be certain that these networks translate in neural cell types. The cell-type and sublocalization specificity of FMRP-targeted mRNAs can suggest that FMRP is likely to form unique complexes in different cell types and within different cellular compartments. To our knowledge, only a few FMRP interactome studies have been conducted in neuronal cells, and while these studies are limited, understanding FMRP-interacting protein networks could answer key questions for the FXS field. If FMRP-interacting proteins are unique across different neural cell types, could this differing pattern of expression be a contributing factor to the phenotypic diversity in individuals with FXS, and in turn clarify the subgroup responses in prior trials of targeted therapies for individuals with FXS? A study published recently by Shen et al., examined the FMRP interactome by performing co-immunoprecipitation (CoIP) followed by MS in human cortical neurons differentiated from FMRP-FLAG human pluripotent stem cells (hPSCs). This study revealed 56 FMRP interactors which were enriched for proteins involved in regulation of translation, stress granule assembly, and RNA metabolism (Shen et al., 2023). Consistent with the HeLa FMRP interactome (Taha et al., 2021), UBAP2, DDX1, and DDX3X were found to interact with FMRP in human cortical neurons (Shen et al., 2023). The group noted that their CoIP data correlated to their CLIP data in that both demonstrated 1) a strong enrichment for genes that presented with lower expression during postnatal development than prenatal development (referred as genes with “falling trajectories), and 2) enrichment for sequences-constrained genes, with 20 of 36 CLIP targets and 28 of 56 CoIP interactors reporting high probability of loss-of-function intolerance scores, which is consistent with the notion that highly constrained genes are intolerant to haploinsufficiency and coordinate critical functions during prenatal development (Shen et al., 2023). Specifically, this study focused on the FMRP interactor CNOT1, revealing that FMRP-CNOT1 interaction is required to maintain mRNA levels of receptor for activated C kinase 1 (RACK1), a human specific FMRP mRNA target. Additionally, the study demonstrated the role of RACK1 in maintaining mitochondrial function, suggesting that reduced RACK1 expression is a contributing factor to mitochondrial defects observed in FXS (Shen et al., 2023). Though not a human model study, Kieffer et al., aimed to define nuclear FMRP interacting proteins to complement the increasing evidence that FMRP is involved in nuclear processes besides its association with nucleocytoplasmic RNA export. The group conducted GST-pulldown and MS on forebrain nuclei isolated from postnatal day 14 rats and focused on proteins that bind to the N-terminus of FMRP due to the N-terminal location of the nuclear localization signal of FMRP. While FMRP also contains a centrally localized nuclear export signal, this study did not investigate this region for protein interactions since the majority of FMRP-interacting proteins bind to C- and N-terminal regions of FMRP. This study revealed 55 FMRP interacting proteins, four of which were previously reported to be associated with FMRP such as Fragile X Related Protein 1 (FXRIP) (Zhang et al., 1995), the zinc finger RNA binding protein (ZFR) (Worringer et al., 2009), the splicing factor Rbm14 (Zhou et al., 2017), and Adar, the mRNA editing enzyme (Shamay-Ramot et al., 2015) (Kieffer et al., 2022). Three novel protein interactors were validated though immunoblot analysis and confocal microscopy: (1) Ddx41, a multi-functional DEAD box helicase involved in pre-mRNA splicing and genome stability; (2) Poldip3 (polymerase δ-interacting protein), a protein involved in cellular DNA replication, nuclear mRNA export, and functions in the exon-exon junction complex by recruiting the SK6 kinase on newly synthesized mRNAs; and (3) Hnrnpa3 (heterogeneous nuclear ribonucleoprotein A3), a protein functions in telomer maintenance in the nucleus and in mRNA splicing, stability, and trafficking of mRNAs in the cytoplasm (Kieffer et al., 2022). Two interesting observations came from this study regarding previously known FMRP-interacting proteins. This study did not identify the nucleocytoplasmic shuttling protein, Nuclear FMRP Interacting Protein 1 (NuFIP1), which is known to interact with the N-terminal domain of FMRP (Bardoni et al., 1999). Furthermore, the identification of FXRIP as a nuclear FMRP-interacting protein is novel in the sense that this protein is known to interact with FMRP in the cytoplasm, presenting a potentially unknown function for this protein (Kieffer et al., 2022). The 55 proteins identified in this study were compared to the 58 FMRP-interacting proteins identified in Shen et al., study on human cortical neurons, and while there were no consistencies between the nuclear rat forebrain and human cortical neuron interactomes, this can be explained by distinct experimental conditions. However, there were four shared proteins between the rat forebrain interactome and HeLa cell interactome described in Taha et al., which were FXRIP, the chromatin remodeling factor CHD4, and the transcription factors TCF20 and ZNF638 (Taha et al., 2021). The minimal overlap between all four interactome studies could be expected due to the functional nature of FMRP. Since FMRP mRNA targets are cell type specific and developmentally regulated, one can speculate that the proteins which guide and facilitate FMRP would also exhibit similar patterns. The data presented in these studies considerably expand functional candidate protein networks of FMRP and suggests that FMRP functions in many fundamental cellular processes throughout the body beyond the nervous system.
3.3. Multiomic approach to studying fragile X syndrome: Transcriptome and proteome analysis
A few human model studies on FXS have taken an integrative multiomic approach by simultaneously conducting transcriptomic and proteomic studies. Singularly, analysis of global mRNA or protein changes offers insight to which biological pathways/processes are dysregulated and both are very useful for drawing correlations to disease; however, single-omic approaches are limited to reflecting reactive processes rather than causative processes (Hasin et al., 2017). Integrating a multiomic approach provides the potential to elucidate causative changes that define diseases and can provide researchers the resolving-power to establish relationships between molecular signatures and phenotypic manifestations.
Utami, KH. et al. performed genome-wide transcriptome and proteome profiling on an isogenic human pluripotent stem cell (hPSCs) model of FXS and FXS neurons differentiated from WCMC-37 FXS cells (FXS hPSCs). FMR1 KO neurons were generated by using CRISPR/Cas9 to introduce indels in exon 3 of FMR1. RNA-seq was performed on day 37 control neurons, FMR1 KO neurons, and FXS neurons (n = 4 per genotype). An overlap of 3110 differently expressed genes from FXS and FMR1 KO neurons were altered compared with control neurons, in which 1525 genes were downregulated and 1585 were upregulated (Utami et al., 2020). Functional annotation of the differentially expressed genes showed enrichment of gene ontology terms including neuron differentiation and neurodevelopment, neurogenesis, and neurotransmission for downregulated genes, whereas RNA processing and transport, translation, and cell cycle processes were the most enriched for upregulated genes (Utami et al., 2020). Three genes involved in neuron differentiation, DSCAM, GAP43, and PTPRT were validated using qRT-PCR(Utami et al., 2020). The group subsequently performed MS proteomics on day 37 control neurons, FMR1 KO neurons, and FXS neurons, which identified a total of 4210 proteins after data filtering. FMR1 KO cells presented with a considerably higher number of differential proteins compared to FXS neurons with 1247 proteins showing downregulation and 951 proteins showing upregulation in the FMR1 KO cells (Utami et al., 2020). In FXS neurons, 343 proteins were downregulated, whereas 234 proteins were upregulated. Gene ontology terms for the differential proteins in the FMR1 KO neurons showered similar categories as the RNA-seq data; however, the differential proteins in the FXS neuron showed enrichment in vesicle transport and synaptic signaling, cell cycle processes, DNA replication, and DNA metabolism. For example, CNTNAP2, GPRIN3, KIF5c, and CNTN1 were all found downregulated in FXS neurons and have previously been associated with FXS and other neurodevelopmental disorders (Utami et al., 2020). Though there were apparent differences between the FMR1 KO and FXS neurons, 244 downregulated proteins and 141 upregulated proteins shared common changes between the two cells line (Utami et al., 2020). The cellular processes related to downregulated proteins were neurotransmitter secretion, synaptic transmission, and dendrite development whereas upregulated proteins were related to DNA replication and cell cycle processes. An important aspect to note is that the data presented many categories like those identified in the transcriptional enrichment analysis, highlighting that proteomic and transcriptomic analyses complement each other and that the RNA level translated to the protein level, thus providing another layer of functional insight.
Kurosaki, T. et al. applied an integrative omics approach to human FMR1 KO SH-SY5Y neuroblastoma cells. Previously, the group reported that knockdown of FMRP resulted in hyperactivated decay rate of mRNAs targeted for nonsense-mediated mRNA decay (NMD), a phenomenon dependent on translation (Kurosaki et al., 2021). Building on their previous findings, the group performed a comprehensive study utilizing SILAC coupled with LC-MS/MS on WT and FMR1 KO SH-SY5Y cells. RNA-seq was simultaneously performed; however, unlike the study conducted by Utami, KH. et al. (Utami et al., 2020), RNA-seq was conducted to normalize the level of individual proteins to the level of mRNA from which protein derives. The group demonstrated that in the absence of FMRP, most mRNAs that are normally transcriptionally regulated by FMRP undergo the largest increase in translation, destabilizing, and decay (Kurosaki et al., 2022). Furthermore, FMRP was shown to stabilize mRNAs and decrease translation by binding to solely the 5’ UTR, coding sequences, or 3’ UTRs via structed GC-rich sequences (Kurosaki et al., 2022), which is actually in disagreement with a study conducted by Shu, H. et al. that noted an essential role of FMRP on codon optimality-dependent mRNA stability (Shu et al., 2020). The results indicate that mRNA expression in FMR1 KO cells is reduced in abundance, but FMR1 KO cells generate more protein per mRNA whether they bind FMRP (Kurosaki et al., 2022). In total, 2160 proteins were upregulated, and 1098 proteins were downregulated in the FMR1 KO cells. Upregulated proteins were noted to be involved in synaptic signaling pathways, glycolysis, and Parkinson’s disease whereas downregulated proteins are enriched for functions in pyridine and cholesterol biosynthesis (Kurosaki et al., 2022). The top five proteins whose abundance was most changed in FMR1 KO cells were selected for WB validation, demonstrating that FMRP loss increased the abundance of chromodomain helicase DNA binding protein 5 (CHD5), Unc-13 homolog D (UNC13D), and rabphilin 3 A (RPH3A), and decreased the abundance of N-myc downstream regulated 1 (NDRG1), transcriptional mediator complex subunit 1 (MED1), and post-glycosylphosphatidylinositol attachment to proteins inositol deacylase 1 (PGAP1) (Kurosaki et al., 2022). The group also conducted anti-FMRP CoIPs followed by LC-MS/MS and identified the major cytoplasmic poly (A)-binding protein, PABPC1, as a direct interactor with FMRP. By quantitating the abundance of poly(A), the group confirmed that efficient FMRP binding to mRNAs required a poly(A) tail and protects mRNAs from deadenylation (Kurosaki et al., 2022). This finding is consistent with the functional role of FMRP in maintaining mRNA stability.
3.4. Analysis of human biofluids in patients with fragile X syndrome
The use of human biofluids, such as blood, urine, and cerebrospinal fluid has become an invaluable resource in proteomics research for the study of various diseases. Biofluid-based proteomics offers a non-invasive or minimally invasive approach to access valuable molecular information underlying disease. In the context of FXS, these studies have profound implications for monitoring treatment responses, understanding the heterogeneity seen across patients, and may open avenues to the development of targeted and personalized therapeutic approaches. While biofluid-based proteomics in FXS are relatively lacking, a few studies have utilized plasma and peripheral blood mononuclear cells (PBMCs) to analyze these subproteomes in patients with FXS.
Dionne and Corbin conducted a proteomics study aimed to address the current lack of diagnostic tools to assess patient prognosis upon diagnosis. To address this, PBMCs isolated from blood samples collected from seven male FXS patients and seven age-matched controls were used to study the nascent proteome by employing BONCAT method combined with label-free mass spectrometry to identify and quantify the newly synthesized proteins in PBMCs (Dionne and Corbin, 2021). The patients chosen for the study all presented with fully methylated FMR1 alleles on the basis that the inclusion of mosaic or female patients, both of which express variable FMRP levels, would increase the heterogeneity within the FXS cohort (Dionne and Corbin, 2021). Notably, the researchers also used PBMCs from the same experimental groups to analyze the total proteome; however, the focus on biomarker identification was directed towards the nascent proteome since the loss of FMRP leads to aberrant protein synthesis (Dionne and Corbin, 2021). Analysis of the total proteome revealed 1770 proteins of which 200 were found to be dysregulated in FXS samples (135 upregulated proteins, 65 downregulated proteins). The nascent proteome revealed 30 dysregulated proteins among the 109 identified, of which 17 were upregulated and 13 downregulated. Additionally, of the 30 dysregulated proteins, 20 were noted to be mRNA bound and translationally regulated by FMRP. The researchers focused on 11 proteins that revealed to be dysregulated using both approaches. Consistent between both approaches, TLN1, FERMT3, HIST1H4A, ILK, MPO, and VCL were upregulated, while AHNAK was downregulated; however, APT2A3, PDIA3, PF4, and ANXA2 presented with opposite expression levels between the nascent and total proteome, suggesting this could arise from alterations in their turnover rates (Dionne and Corbin, 2021).
A previously mentioned study conducted by Bowling, H. et al. (Bowling et al., 2019), which utilized an optimized adaptation of the BONCAT technique (Bowling et al., 2020) to identify newly synthesis proteins in Fmr1 KO and WT mice hippocampus under basal and stimulated condition, also screened human plasma from 11 patients with FXS against healthy controls based. Seven candidate proteins identified in their BONLAC screens were chosen for validation by Western blotting in plasma samples based on the top GO categories of biosynthesis proteins, cell membrane proteins, intracellular signaling proteins, and synapse proteins: ACO2, HK1, BDNF, GRIN2B, mGluR5, and SYNGAP. Of the seven proteins, three proteins (HK1, RAS, and ACO2) exhibited a clear difference in abundance between FXS patient plasma and control plasma. Notabaly, RAS and ACO2 were significantly downregulated while HK1 presented with a downregulated trend.
A point worth mentioning is that there were no consistencies in the nascent proteome changes between the studies conducted by Dionne and Corbin (Dionne and Corbin, 2021) and Bowling, H. et al. (Bowling et al., 2019); however, this could be explained by the differences in protein composition between PBMCs and the mouse hippocampus. Furthermore, the limited number of nascent proteins identified in PBMCs compared to the number of nascent proteins identified in mouse hippocampus could be attributed to sample preparation. In the study conducted by Dionne and Corbin (Dionne and Corbin, 2021), PBMCs were labeled with AHA for two hours, limiting their analysis to proteins with high turnover rates whereas Bowling, H. et al. (Bowling et al., 2019) labeled their samples overnight.
4. Conclusions and future outlook
Here we present a comprehensive summary of MS-based proteomic studies conducted on FXS. Proteomics has offered a multidimensional view of FXS through unraveling intricate molecular networks and pointing towards novel avenues for therapeutic interventions. Understanding the proteomic landscape of FXS not only enhances our comprehension of FXS, but also provides hope for the development of precision therapies designed to improve the lives of individuals affected by FXS and related neurodevelopmental conditions. Although the field has made significant progress in characterizing the fragile x proteome, there are gaps in the literature that should be considered for future investigation. The analysis of human biomaterials such as blood serum or urine can reveal protein biomarkers useful to clinical applications such as treatment monitoring. A few studies have characterized the proteome of blood samples in patients with FXS; however, these studies primarily focused on nascent proteome changes. Additionally, the FXS field would substantially benefit from additional proteomic studies on human FXS neurons and subtype neural cells such as astrocytes. Although in early stages of implementation, MS serves as a dynamic technique that has the great potential in furthering the general understanding of FXS.
Acknowledgments
Figures were created using Biorender.com.
Funding
This work was supported by Rush University Medical Center, University of Illinois Chicago, University of Wisconsin-Madison and grants to XZ (NIH R01MH118827, R01MH116582, DOD IIRA grant W81XWH-22–1-0621) and SMC (NIH R01NS114413, R01NS124784, U01FD008126).
Footnotes
CRediT authorship contribution statement
Diana A. Abbasi: Writing – review & editing, Writing – original draft, Investigation, Formal analysis, Conceptualization. Elizabeth Berry-Kravis: Writing – review & editing, Supervision, Formal analysis. Xinyu Zhao: Writing – review & editing, Supervision. Stephanie M. Cologna: Writing – review & editing, Supervision.
Declaration of competing interest
The authors declare that were no competing interests associated with the manuscript.
Data availability
No data was used for the research described in the article.
References
- Aloisi E, et al. , 2017. Altered surface mGluR5 dynamics provoke synaptic NMDAR dysfunction and cognitive defects in Fmr1 knockout mice. Nat. Commun. 8, 1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagni C, et al. , 2012. Fragile X syndrome: causes, diagnosis, mechanisms, and therapeutics. J. Clin. Invest. 122, 4314–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai F, Witzmann FA, 2007. Synaptosome proteomics. Subcell. Biochem. 43, 77–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardoni B, et al. , 1999. A novel RNA-binding nuclear protein that interacts with the fragile X mental retardation (FMR1) protein. Hum. Mol. Genet. 8, 2557–2566. [DOI] [PubMed] [Google Scholar]
- Bardoni B, et al. , 2003. 82-FIP, a novel FMRP (fragile X mental retardation protein) interacting protein, shows a cell cycle-dependent intracellular localization. Hum. Mol. Genet. 12, 1689–1698. [DOI] [PubMed] [Google Scholar]
- Bartley CM, et al. , 2016. Mammalian FMRP S499 is phosphorylated by CK2 and promotes secondary phosphorylation of FMRP. eNeuro 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry-Kravis E, 2014. Mechanism-based treatments in neurodevelopmental disorders: fragile X syndrome. Pediatr. Neurol. 50, 297–302. [DOI] [PubMed] [Google Scholar]
- Berry-Kravis E, et al. , 2011. Targeted treatments for fragile X syndrome. J. Neurodev. Disord. 3, 193–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling H, et al. , 2016. BONLAC: a combinatorial proteomic technique to measure stimulus-induced translational profiles in brain slices. Neuropharmacology 100, 76–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling H, et al. , 2019. Altered steady state and activity-dependent de novo protein expression in fragile X syndrome. Nat. Commun. 10, 1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling HL, et al. , 2020. Optimization of protocols for detection of De novo protein synthesis in whole blood samples via Azide-alkyne cycloaddition. J. Proteome Res. 19, 3856–3866. [DOI] [PubMed] [Google Scholar]
- Broek JAC, et al. , 2016. Synaptic vesicle dynamic changes in a model of fragile X. Mol. Autism. 7, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bülow P, et al. , 2019. Homeostatic intrinsic plasticity is functionally altered in Fmr1 KO cortical neurons. Cell Rep. 26, 1378–1388.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bülow P, et al. , 2021. FMRP attenuates activity dependent modifications in the mitochondrial proteome. Mol. Brain 14, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carotenuto M, et al. , 2019. Polysomnographic findings in fragile X syndrome children with EEG abnormalities. Behav. Neurol. 2019, 5202808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceman S, et al. , 2003. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum. Mol. Genet. 12, 3295–3305. [DOI] [PubMed] [Google Scholar]
- Chen L, et al. , 2011. High-resolution methylation polymerase chain reaction for fragile X analysis: evidence for novel FMR1 methylation patterns undetected in southern blot analyses. Genet. Med. 13, 528–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie SB, et al. , 2009. The FXG: a presynaptic fragile X granule expressed in a subset of developing brain circuits. J. Neurosci. 29, 1514–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlhaus R, 2018. Of men and mice: modeling the fragile X syndrome. Front. Mol. Neurosci. 11, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai SH, et al. , 2014. Activation of mGluR5 attenuates NMDA-induced neurotoxicity through disruption of the NMDAR-PSD-95 complex and preservation of mitochondrial function in differentiated PC12 cells. Int. J. Mol. Sci. 15, 10892–10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, et al. , 2011. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne O, Corbin F, 2021. A new strategy to uncover fragile X proteomic biomarkers using the nascent proteome of peripheral blood mononuclear cells (PBMCs). Sci. Rep. 11, 15148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el Bekay R, et al. , 2007. Enhanced markers of oxidative stress, altered antioxidants and NADPH-oxidase activation in brains from fragile X mental retardation 1-deficient mice, a pathological model for fragile X syndrome. Eur. J. Neurosci. 26, 3169–3180. [DOI] [PubMed] [Google Scholar]
- Ge SX, et al. , 2020. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36, 2628–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godler DE, et al. , 2010. Methylation of novel markers of fragile X alleles is inversely correlated with FMRP expression and FMR1 activation ratio. Hum. Mol. Genet. 19, 1618–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godler DE, et al. , 2011. FMR1 intron 1 methylation predicts FMRP expression in blood of female carriers of expanded FMR1 alleles. J. Mol. Diagn. 13, 528–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godler DE, et al. , 2012. Fragile X mental retardation 1 (FMR1) intron 1 methylation in blood predicts verbal cognitive impairment in female carriers of expanded FMR1 alleles: evidence from a pilot study. Clin. Chem. 58, 590–598. [DOI] [PubMed] [Google Scholar]
- Goldsmith CS, Bell-Pedersen D, 2013. Diverse roles for MAPK signaling in circadian clocks. Adv. Genet. 84, 1–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt EJ, Spradling AC, 2018. Fragile X mental retardation 1 gene enhances the translation of large autism-related proteins. Science 361, 709–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, et al. , 2021. FMRP regulates mRNAs encoding distinct functions in the cell body and dendrites of CA1 pyramidal neurons. Elife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin Y, et al. , 2017. Multi-omics approaches to disease. Genome Biol. 18, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber KM, et al. , 2001. Chemical induction of mGluR5- and protein synthesis–dependent long-term depression in hippocampal area CA1. J. Neurophysiol. 86, 321–325. [DOI] [PubMed] [Google Scholar]
- Jacquemont S, et al. , 2018. Protein synthesis levels are increased in a subset of individuals with fragile X syndrome. Hum. Mol. Genet. 27, 2039–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin DZ, et al. , 2015. Metabotropic glutamate receptor 5 upregulates surface NMDA receptor expression in striatal neurons via CaMKII. Brain Res. 1624, 414–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Anderson P, 2009. Regulation of translation by stress granules and processing bodies. Prog. Mol. Biol. Transl. Sci. 90, 155–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer F, et al. , 2022. Combining affinity purification and mass spectrometry to define the network of the nuclear proteins interacting with the N-terminal region of FMRP. Front. Mol. Biosci. 9, 954087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SW, Cho KJ, 2014. Activity-dependent alterations in the sensitivity to BDNF-TrkB signaling may promote excessive dendritic arborization and spinogenesis in fragile X syndrome in order to compensate for compromised postsynaptic activity. Med. Hypotheses 83, 429–435. [DOI] [PubMed] [Google Scholar]
- Klemmer P, et al. , 2011. Proteomics, ultrastructure, and physiology of hippocampal synapses in a fragile X syndrome mouse model reveal presynaptic phenotype. J. Biol. Chem. 286, 25495–25504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko ML, et al. , 2013. Circadian phase-dependent effect of nitric oxide on L-type voltage-gated calcium channels in avian cone photoreceptors. J. Neurochem. 127, 314–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T, et al. , 2021. NMD abnormalities during brain development in the Fmr1-knockout mouse model of fragile X syndrome. Genome Biol. 22, 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T, et al. , 2022. Integrative omics indicate FMRP sequesters mRNA from translation and deadenylation in human neuronal cells. Mol. Cell 82, 4564–4581.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzniewska B, et al. , 2020. Mitochondrial protein biogenesis in the synapse is supported by local translation. EMBO Rep. 21, e48882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumonnier F, et al. , 2006. Association of a functional deficit of the BKCa channel, a synaptic regulator of neuronal excitability, with autism and mental retardation. Am. J. Psychiatry 163, 1622–1629. [DOI] [PubMed] [Google Scholar]
- Lessmann V, et al. , 1994. BDNF and NT-4/5 enhance glutamatergic synaptic transmission in cultured hippocampal neurones. Neuroreport 6, 21–25. [DOI] [PubMed] [Google Scholar]
- Li M, et al. , 2020. Identification of FMR1-regulated molecular networks in human neurodevelopment. Genome Res. 30, 361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao L, et al. , 2008. Quantitative proteomic analysis of primary neurons reveals diverse changes in synaptic protein content in fmr1 knockout mice. Proc. Natl. Acad. Sci. USA. 105, 15281–15286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licznerski P, et al. , 2020. ATP synthase c-subunit leak causes aberrant cellular metabolism in fragile X syndrome. Cell 182, 1170–1185.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano R, et al. , 2014. Fragile X spectrum disorders. Intract. Rare Dis. Res. 3, 134–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, et al. , 2010. Fragile x mental retardation protein regulates proliferation and differentiation of adult neural stem/progenitor cells. PLoS Genet. 6, e1000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, et al. , 2024. On-tissue spatial proteomics integrating MALDI-MS imaging with shotgun proteomics reveals soy consumption-induced biomarkers in a fragile X syndrome mouse model. ACS Chem Neurosci 15 (1), 119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macklin A, et al. , 2020. Recent advances in mass spectrometry based clinical proteomics: applications to cancer research. Clin. Proteomics 17, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matic K, et al. , 2014. Quantitative phosphoproteomics of murine Fmr1-KO cell lines provides new insights into FMRP-dependent signal transduction mechanisms. J. Proteome Res. 13, 4388–4397. [DOI] [PubMed] [Google Scholar]
- Maurin T, et al. , 2018. HITS-CLIP in various brain areas reveals new targets and new modalities of RNA binding by fragile X mental retardation protein. Nucleic Acids Res. 46, 6344–6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monzo K, et al. , 2010. Proteomic analysis reveals CCT is a target of fragile X mental retardation protein regulation in Drosophila. Dev. Biol. 340, 408–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita T, et al. , 2019. Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 20, 156–174. [DOI] [PubMed] [Google Scholar]
- Ngounou Wetie AG, et al. , 2014. Mass spectrometry for the study of autism and neurodevelopmental disorders. Adv. Exp. Med. Biol. 806, 525–544. [DOI] [PubMed] [Google Scholar]
- Pal R, et al. , 2015. The potential of proteomics in understanding neurodegeneration. Int. Rev. Neurobiol. 121, 25–58. [DOI] [PubMed] [Google Scholar]
- Qiu W, et al. , 2020. Phosphopeptide enrichment for phosphoproteomic analysis – a tutorial and review of novel materials. Anal. Chim. Acta 1129, 158–180. [DOI] [PubMed] [Google Scholar]
- Rath S, et al. , 2021. MitoCarta3.0: an updated mitochondrial proteome now with suborganelle localization and pathway annotations. Nucleic Acids Res. 49, D1541–d1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes ST, et al. , 2020. GABA measurement in a neonatal fragile X syndrome mouse model using (1)H-magnetic resonance spectroscopy and mass spectrometry. Front. Mol. Neurosci. 13, 612685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, Zhao X, 2021. The molecular biology of FMRP: new insights into fragile X syndrome. Nat. Rev. Neurosci. 22, 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcedo-Arellano MJ, et al. , 2020. Fragile X syndrome and associated disorders: clinical aspects and pathology. Neurobiol. Dis. 136, 104740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro MR, et al. , 2012. Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Annu. Rev. Pathol. 7, 219–245. [DOI] [PubMed] [Google Scholar]
- Sawicka K, et al. , 2019. FMRP has a cell-type-specific role in CA1 pyramidal neurons to regulate autism-related transcripts and circadian memory. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo SS, et al. , 2022. Excess ribosomal protein production unbalances translation in a model of fragile X syndrome. Nat. Commun. 13, 3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethna F, et al. , 2014. From FMRP function to potential therapies for fragile X syndrome. Neurochem. Res. 39, 1016–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamay-Ramot A, et al. , 2015. Fmrp interacts with Adar and Regulates RNA editing, synaptic density and locomotor activity in zebrafish. PLoS Genet. 11, e1005702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M, et al. , 2023. Species-specific FMRP regulation of RACK1 is critical for prenatal cortical development. Neuron 111 (24), 3988–4005 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu H, et al. , 2020. FMRP links optimal codons to mRNA stability in neurons. Proc. Natl. Acad. Sci. USA 117, 30400–30411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, et al. , 1995. FXR1, an autosomal homolog of the fragile X mental retardation gene. EMBO J. 14, 2401–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha MS, et al. , 2021. Novel FMRP interaction networks linked to cellular stress. FEBS J. 288, 837–860. [DOI] [PubMed] [Google Scholar]
- Talvio K, et al. , 2021. Increased iron content in the heart of the Fmr1 knockout mouse. Biometals 34, 947–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B, et al. , 2015. Fmr1 deficiency promotes age-dependent alterations in the cortical synaptic proteome. Proc. Natl. Acad. Sci. USA 112, E4697–E4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telias M, 2019. Molecular mechanisms of synaptic dysregulation in fragile X syndrome and autism Spectrum disorders. Front. Mol. Neurosci. 12, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till SM, et al. , 2011. A presynaptic role for FMRP during protein synthesis-dependent long-term plasticity in Aplysia. Learn. Mem. 18, 39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utami KH, et al. , 2020. Integrative analysis identifies key molecular signatures underlying neurodevelopmental deficits in fragile X syndrome. Biol. Psychiatry 88, 500–511. [DOI] [PubMed] [Google Scholar]
- Verkerk AJ, et al. , 1991. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65, 905–914. [DOI] [PubMed] [Google Scholar]
- Witze ES, et al. , 2007. Mapping protein post-translational modifications with mass spectrometry. Nat. Methods 4, 798–806. [DOI] [PubMed] [Google Scholar]
- Woods AG, et al. , 2015. Autism spectrum disorder: an omics perspective. Proteomics Clin. Appl. 9, 159–168. [DOI] [PubMed] [Google Scholar]
- Wormwood KL, et al. , 2019. Mass spectrometry for the study of autism and neurodevelopmental disorders. Adv. Exp. Med. Biol. 1140, 477–499. [DOI] [PubMed] [Google Scholar]
- Worringer KA, et al. , 2009. The zinc finger protein Zn72D and DEAD box helicase belle interact and control maleless mRNA and protein levels. BMC Mol. Biol. 10, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YY, et al. , 2022. Lysine acetylome profiling in mouse hippocampus and its alterations upon FMRP deficiency linked to abnormal energy metabolism. J. Proteome 269, 104720. [DOI] [PubMed] [Google Scholar]
- Xu B, et al. , 2018. Proteomic profiling of brain and testis reveals the diverse changes in ribosomal proteins in fmr1 knockout mice. Neuroscience 371, 469–483. [DOI] [PubMed] [Google Scholar]
- Zhang G, Neubert TA, 2009. Use of stable isotope labeling by amino acids in cell culture (SILAC) for phosphotyrosine protein identification and quantitation. Methods Mol. Biol. 527 (79–92), xi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, et al. , 1995. The fragile X mental retardation syndrome protein interacts with novel homologs FXR1 and FXR2. EMBO J. 14, 5358–5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YQ, et al. , 2001. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell 107, 591–603. [DOI] [PubMed] [Google Scholar]
- Zhang YQ, et al. , 2004. The Drosophila fragile X-related gene regulates axoneme differentiation during spermatogenesis. Dev. Biol. 270, 290–307. [DOI] [PubMed] [Google Scholar]
- Zhang YQ, et al. , 2005. Protein expression profiling of the drosophila fragile X mutant brain reveals up-regulation of monoamine synthesis. Mol. Cell. Proteomics 4, 278–290. [DOI] [PubMed] [Google Scholar]
- Zhang Y, et al. , 2014. Dendritic channelopathies contribute to neocortical and sensory hyperexcitability in Fmr1( /y) mice. Nat. Neurosci. 17, 1701–1709. [DOI] [PubMed] [Google Scholar]
- Zhang Z, et al. , 2016. Interconversion of peptide mass spectral libraries Derivatized with iTRAQ or TMT labels. J. Proteome Res. 15, 3180–3187. [DOI] [PubMed] [Google Scholar]
- Zhang J, et al. , 2019. Expression and characterization of human fragile X mental retardation protein isoforms and interacting proteins in human cells. Proteom. Insights. 10, 1178641818825268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou LT, et al. , 2017. A novel role of fragile X mental retardation protein in pre-mRNA alternative splicing through RNA-binding protein 14. Neuroscience 349, 64–75. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.