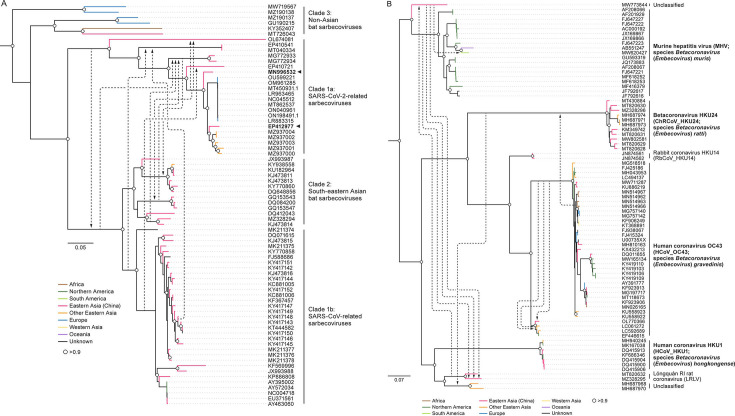
Fig 2.
The ancestry of recombination events in genus Betacoronavirus. (A) Interclade recombination events in Sarbecovirus, the subgenus with the highest number of events. Sarbecoviruses are commonly divided into clades, thus sequences (identified via GenBank accession numbers) are classified accordingly. (B) Interspecies recombination events in Embecovirus, the subgenus with the second-highest number of events. Each arrow represents a recombination event. Arrows in both subfigures represent individual recombination events. The base of an arrow is at the minor parental ancestor (donor of recombination fragment), and the head of the arrow points to the ancestor (or sequence) considered for recombination. The names and abbreviations of officially classified viruses are emphasized in bold print (2). Trees were midpoint-rooted, and directionality of recombination (arrows) is given from the results of the receptor-binding domain (which specifies recombinants and parents). White circles represent nodes with support values > 0.90.