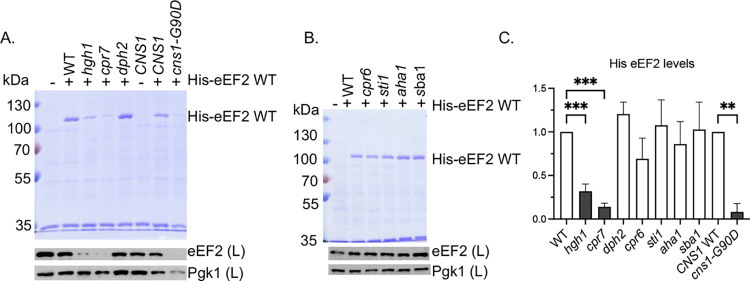
Fig 1. Effect of cochaperone deletion or mutation 6x-His eEF2 levels.
His-eEF2 was isolated from the indicated strain (S1 Table) using nickel resin. Proteins bound to the nickel resin were separated using SDS-PAGE and visualized using stained gels (top). Levels of eEF2 were also detected using immunoblot analysis of whole cell lysates (below). Lane 1: empty vector (YcPlac111). eEF2 levels in the immunoblots in A and B are the combined signal from endogenous eEF2 + plasmid His-eEF2. Anti-Pgk1was used as a loading control. A. Effect of cochaperones previously shown to affect eEF2 folding, as well as a protein (Dph2) required for eEF2 modification. B. Effect of additional Hsp90 cochaperones. C. Quantification of the changes in His-eEF2 levels in panels A and B. The level of His-eEF2 bound to resin in each strain was quantified as described in Materials and Methods. The mean values and standard deviations of three biological replicates, along with a representative of each, are shown. Cochaperones previously linked to eEF2 function are shaded dark gray. Statistical significance was evaluated with GraphPad Prism using Mixed-effects analysis (* P ≤ 0.05; ** P ≤ 0.01; *** P ≤ 0.001). Non-significant values not shown.