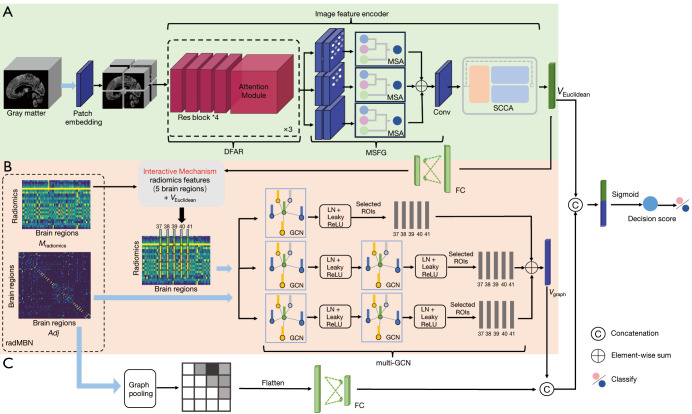
Figure 1.
Architecture and explanation of the proposed MSRNet. (A) The Euclidean representation channel based on the gray-matter volume map. (B) The multi-GCN branch of the graph representation channel based on the radMBN. (C) The fully connected layer branch of the graph representation channel. The interactive mechanism connects the Euclidean and graph representation channels in five disease-related brain regions, as identified in Figure 2. The blue arrows represent the inputs to the model. DFAR, discriminative features by focusing on atrophic regions; MSA, multiscale convolution self-attention module; MSFG, multiscale module that extracts fine-grained information from images; Conv, convolution layer; SCCA, A feature representation enhancement module that integrated spatial attention, channel attention, and coordinate attention; FC, fully connected layers; radMBN, radiomics-based morphology brain network; ROI, region of interest; LN, layer normalization; ReLU, rectified linear unit; Res block, residual block; GCN, graph convolutional neural network; MSRNet, multispatial information representation model; multi-GCN, multilayer graph convolutional neural network.