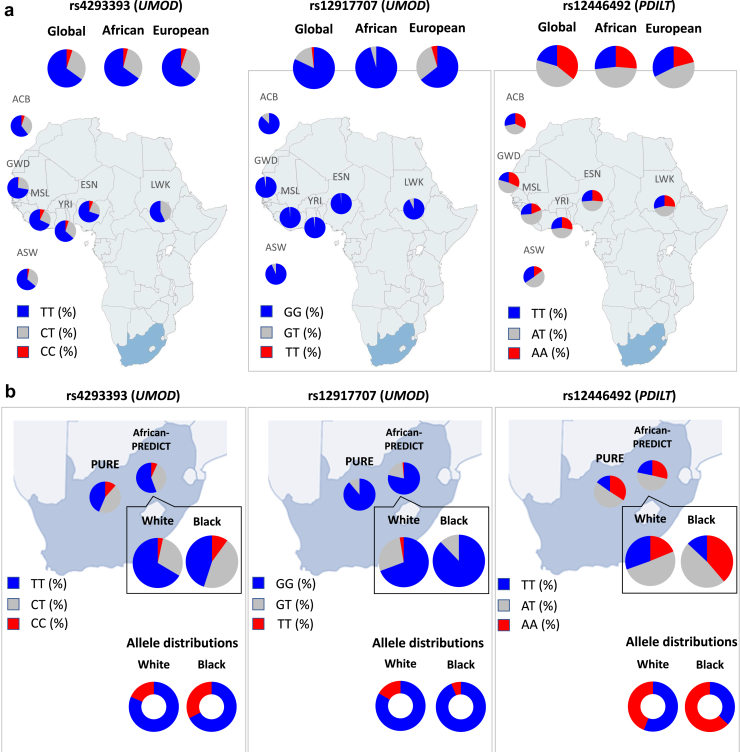
Figure 1.
Distribution of the UMOD/PDILT SNPs in African and European populations from the 1000 Genomes Project, the African-PREDICT and the PURE cohorts. (a) Genotype distribution of rs4293393 (UMOD), rs12917707 (UMOD), and rs12446492 (PDILT) in the African samples from the 1000 Genomes Project, compared to the global and European sample prevalence. (b) Genotype distribution of rs4293393, rs12917707, and rs12446492 in Black and White adults from the African-PREDICT and PURE studies. Blue sections represent the frequencies of “CKD risk” alleles and red sections the frequencies of “CKD protective” alleles. Populations represented in the European ancestry (n = 503) group of the 1000 Genomes Project included: Utah residents from North and West Europe CEU (n = 99), Toscani in Italia TSI (n = 107), Finnish in Finland FIN (n = 99), British in England and Scotland GBR (n = 91), and Iberian population in Spain IBS (n = 107). The African superpopulation is composed of the following populations: African Caribbean in Barbados ACB (n = 96), Gambian in Western Divisions GWD in the Gambia (n = 113), Luhya in Webuye, Kenya LWK (n = 99), Esan in Nigeria ESN (n = 99), Yoruba in Ibadan, Nigeria YRI (n = 108), Mende in Sierra Leone MSL (n = 85), and Americans with African ancestry in Southwest United States ASW (n = 61). The South African population is represented by the North-West Province PURE cohort (n = 1943) and the total of African-PREDICT cohort (n = 1202).