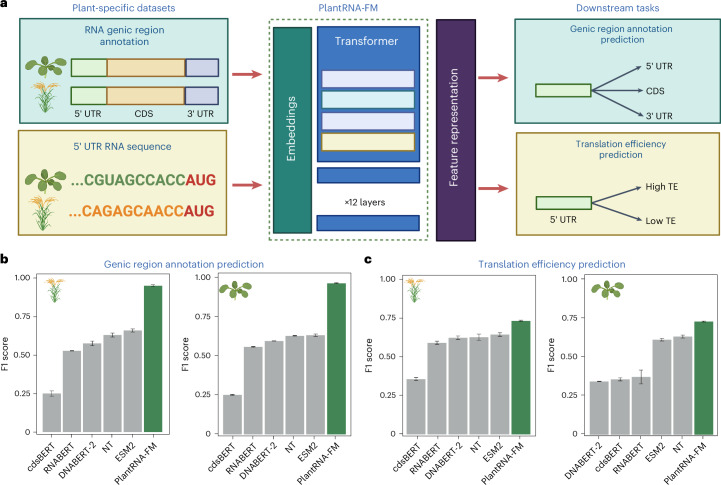
Fig. 2. Fine-tuning PlantRNA-FM on plant-specific datasets.
a, Overview of fine tuning PlantRNA-FM for RNA genic region annotation prediction and RNA TE prediction tasks. A. thaliana and O. sativa were selected as the representative plant species. For the RNA genic region annotation prediction task, RNA sequences from these two species were included, along with three labels: 5′ UTR, CDS and 3′ UTR. For the RNA TE prediction task, the 5′ UTR sequences from these two species were included, along with TE labels (high TE and low TE). b,c, Comparison of the model performance of different pretrained models on RNA genic region annotation prediction and RNA TE prediction tasks. The error bars represent the standard deviation of the F1 scores obtained from three fine-tuning replicates. Panel a was created with BioRender.com.