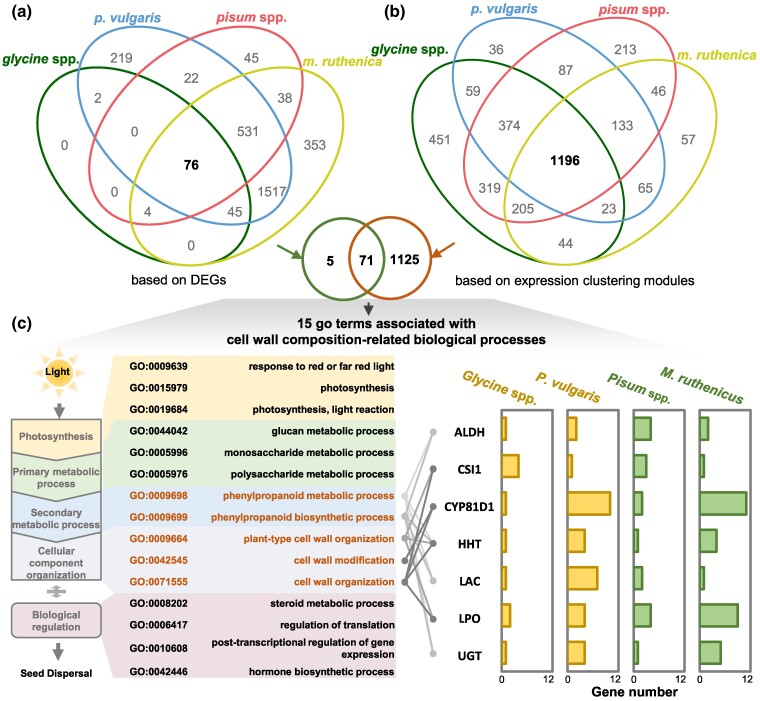
Fig. 3.
Integrative analysis of DEGs and coexpressed genes for legume pod dehiscence. a) Venn of GO enrichment of DEGs. b) Venn of GO enrichment for coexpression modules. c) GO terms related to cell wall composition. Five GO terms and seven structural gene families closely related to pod dehiscence are highlighted, and their gene numbers in DEGs and coexpression modules are given in different legume species. LPO, lactoperoxidase; HHT, ω-hydroxypalmitate O-feruloyl transferase; UGT, UDP-glycosyltransferase; ALDH, aldehyde dehydrogenase; CYP81D1, Cytochrome P450/isoflavone 2′-hydroxylase; LAC, laccase; CSI1, cellulose synthase-interactive protein 1.