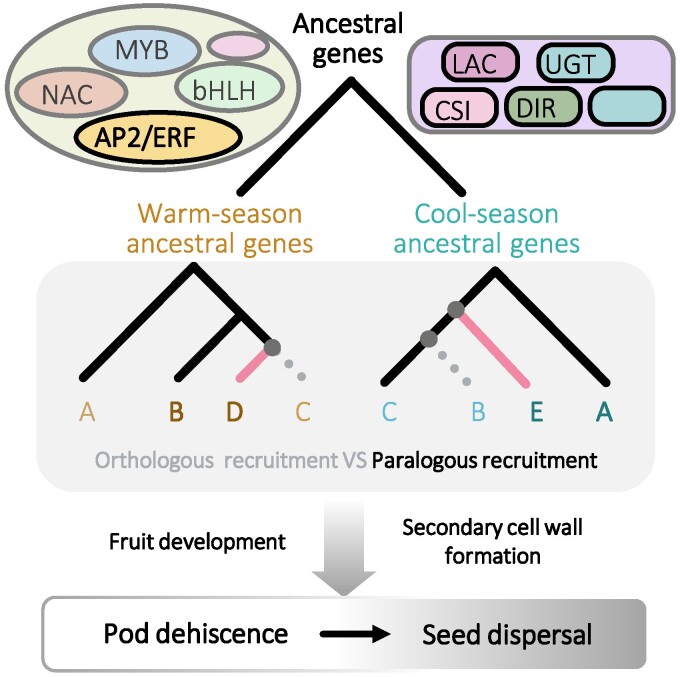
Fig. 7.
Evolution of genetic architecture of pod dehiscence in legumes. Various TF gene families including AP2/ERF-like and structural gene families such as LAC, CSI1, UGT, and DIR might be involved in pod dehiscence formation in legumes. Members in these gene families participate in fruit development and secondary cell wall formation, thus affecting seed dispersal. During evolution, recruiting either orthologous or paralogous members in these gene families to be differentially expressed likely constitutes the genetic architecture and robustness of legume pod dehiscence formation. In legume, paralogous recruitment was the major evolutionary event compared to orthologous recruitment. Orthologous genes are represented by the same letters and paralogous genes are represented by different letters. Moreover, specific gene loss or gene gain might occur, which represented additional evolutionary mechanisms for pod dehiscence variation. Gray dotted line indicates gene loss and pink line represents genes that were specifically originated. CSI1, cellulose synthase-interactive protein 1; DIR, dirigent; LAC, laccase; UGT, UDP-glycosyltransferase.