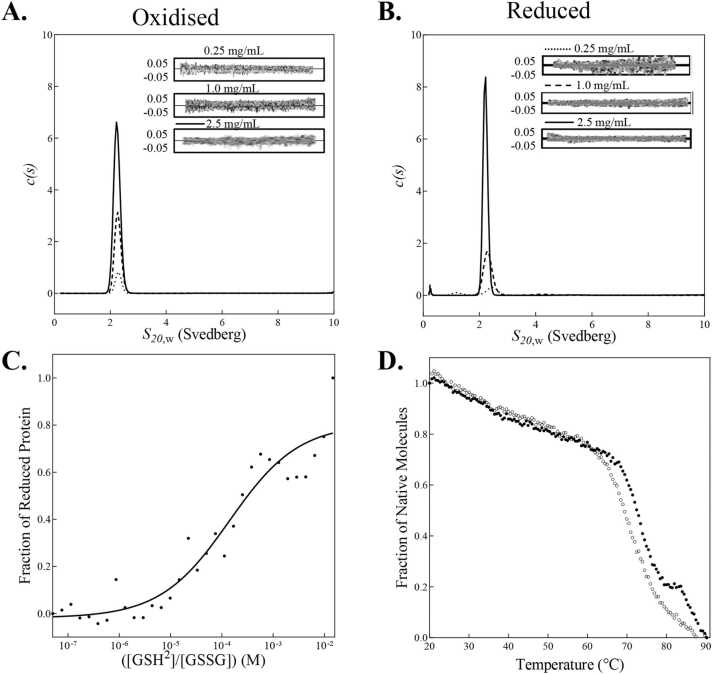
Fig. 3.
Redox Characterisation of FtDsbA1. (A&B.) Overlay of AUC continuous standardised sedimentation coefficient (s20,w) distribution of (A.) oxidised and (B.) reduced FtDsbA1 at 2.5 (solid line), 1 (dashed line) and 0.25 (dotted line) mg/mL. Residuals from the c(s) distribution best fits plotted as a function of radial position from the axis of rotation. (C.) FtDsbA1 redox potential. FtDsbA1 was incubated with various concentrations of reduced glutathione (GSH) overnight and was analysed fluorometrically. The fraction of reduced protein was plotted against log([GSH]²/[GSSG]) to determine the Keq (1.85 ×10⁻⁴ M) and redox potential was then calculated (−129 mV). Data represents one biological replicate with three technical replicates that is representative of three independent replicates. (D.) Thermal melt of oxidised (open circles ○) and reduced (closed circles •) FtDsbA1. CD thermal melts are plotted as a fraction of α-helical content (based on molar ellipticity [θ] at 222 nm) and temperature (°C). Thermal melts represent three technical replicates.