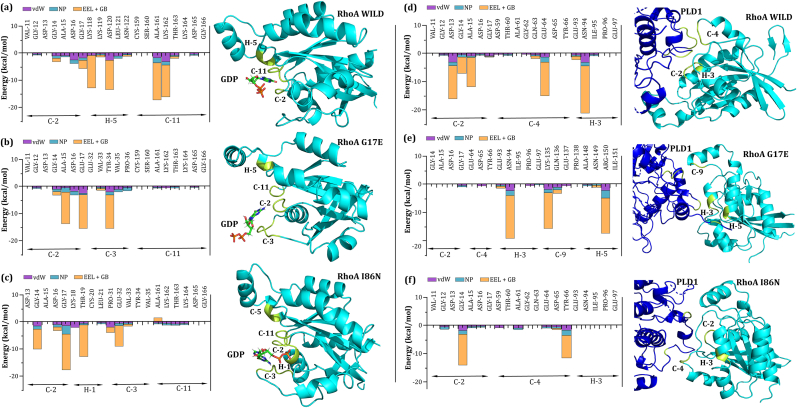
Fig. 10.
Interaction free energy analysis of (a, b, c) Wild-type RhoA, G17E, and I86N mutant residues interacting with GDP, and (d, e, f) Wild-type RhoA, G17E, and I86N mutant residues interacting with PLD1 within a 4 Å binding vicinity. The left panels show the comparison of interaction free energy components (vdW, NP, EEL + GB) for RhoA-GDP complexes, and the right panels display the same for RhoA-PLD1 complexes after 250 ns of molecular dynamics simulations. The most significant residues contributing to the binding free energy in both complexes are highlighted in yellow within the structural representations.