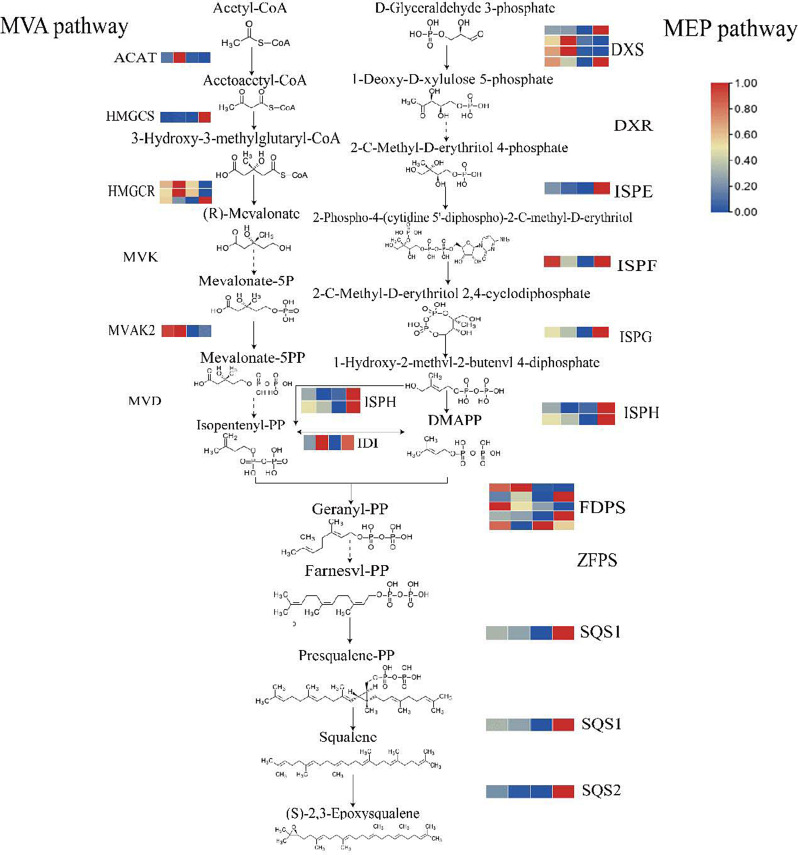
Fig. 4.
Upstream Pathway of Triterpenoid Synthesis in C. paliurus. Solid lines indicate pathways where differential genes have been identified in the transcriptome analysis, while dotted lines represent pathways without identified differential genes. The colored blocks next to each enzyme represent the FPKM (Fragments Per Kilobase of transcript per Million mapped reads) values of the corresponding genes, with expression levels shown from August to November. The color gradient moves from blue (lower expression) to red (higher expression), illustrating the dynamic expression of genes involved in the triterpenoid biosynthesis pathway. Gene name abbreviation: acetyl-CoA acyltransferase (ACAT), 3-hydroxy-3-methylglutaryl-CoA synthetase (HMGS), 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR), mevalonate kinase (MVK), phosphomevalonate kinase (MVAK2), 5-pyrophosphate mevalonate decarboxylase (MVD), 1-deoxyd-xylulose-5-phosphate synthetase (DXS), 1-deoxyd-xylose-5-phosphate reduction isomerase (DXR), 2-C-methyl-D-erythritol 2, 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase (ISPE), 4-cyclodiphosphate synthase (ISPF), (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase (ISPG), 4-hydroxy-3-methylbut-2-en-1-yl diphosphate reductase (ISPH), isopentenyl pyrophosphate isomerase (IDI), farnesyl diphosphate synthase (FDPS), (2Z,6Z)-farnesyl diphosphate synthase (ZFPS), squalene synthase 1 (SQS1), squalene synthase 2 (SQS2)