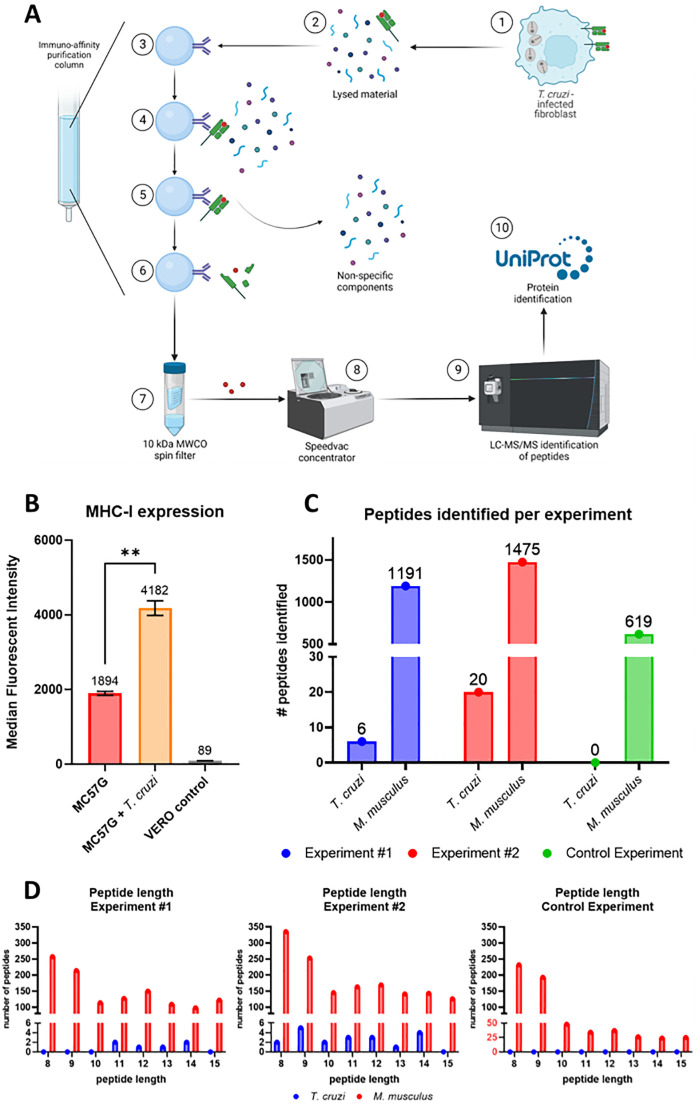
Fig 2. Isolation and identification of the peptides presented on MHC-I of T. cruzi infected MC57G fibroblast cells.
A) Schematic overview of the immunopeptidomics workflow. 1: MC57G murine fibroblasts were co-cultured for 48 hours with T. cruzi Tulahuen trypomastigotes. 2: Infected fibroblasts were harvested and lysed using a non-denaturing lysis buffer. 3: Immuno-affinity purification column was prepared by covalently linking MHC-I–specific mAbs (M1/42) to AminoLink Plus resin. 4: Lysed fibroblast material was loaded on the column, allowing the peptide-loaded MHC-I complexes to bind. 5: Four different wash buffers were used to remove the non-specific components. 6: Acetic acid was used to elute the peptide—MHC-I complexes from the column and dissociate the peptide from the MHC-I. 7: A spin filter column separated the peptide fraction from the MHC-I molecules. 8: Peptides were concentrated using a speedvac concentrator. 9: concentrated peptides were analyzed using LC-MS/MS for their sequence. 10: using the T. cruzi proteome from UniProt T. cruzi proteins were identified. Mouse self-proteins were identified using the C57BL/6J proteome. Figure prepared with Biorender.com. B) T. cruzi infected and non-infected MC57G fibroblasts, as well as green monkey kidney (VERO) cells were stained with anti-mouse MHC-I Alexa Fluor 488. Cells were analyzed by flow cytometry and data was reported as median fluorescent intensity (MFI). Mean and standard deviation are shown and were calculated from six technical replicates. Statistical significance: **: p < 0.01. C) Number of MHC-I binding peptides identified from T. cruzi or mouse for each experiment. D) Distribution of the peptide length for each experiment.