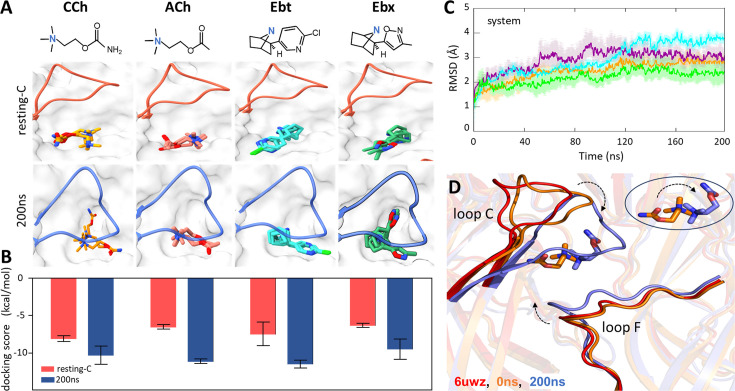
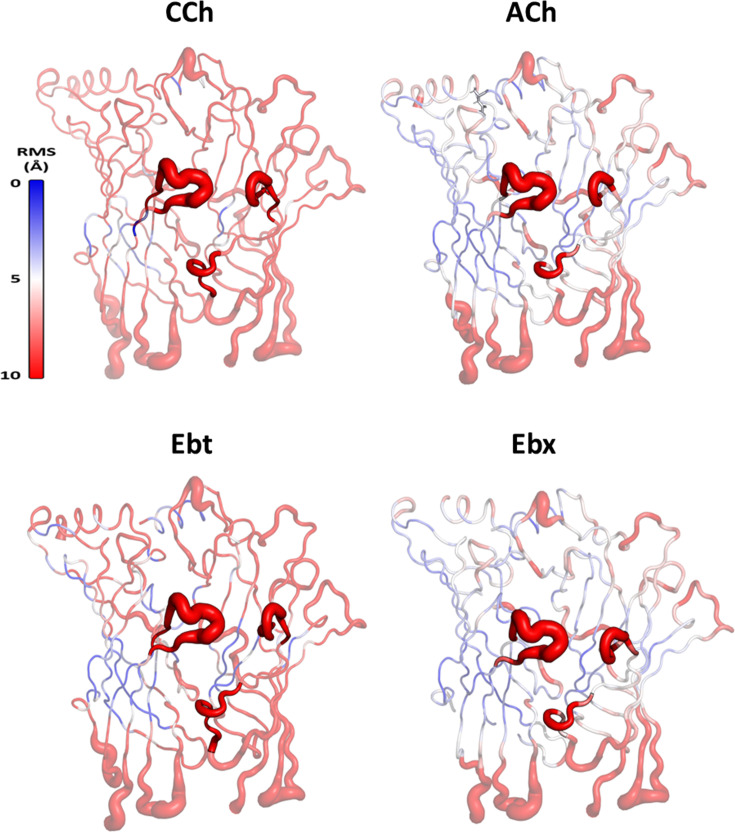
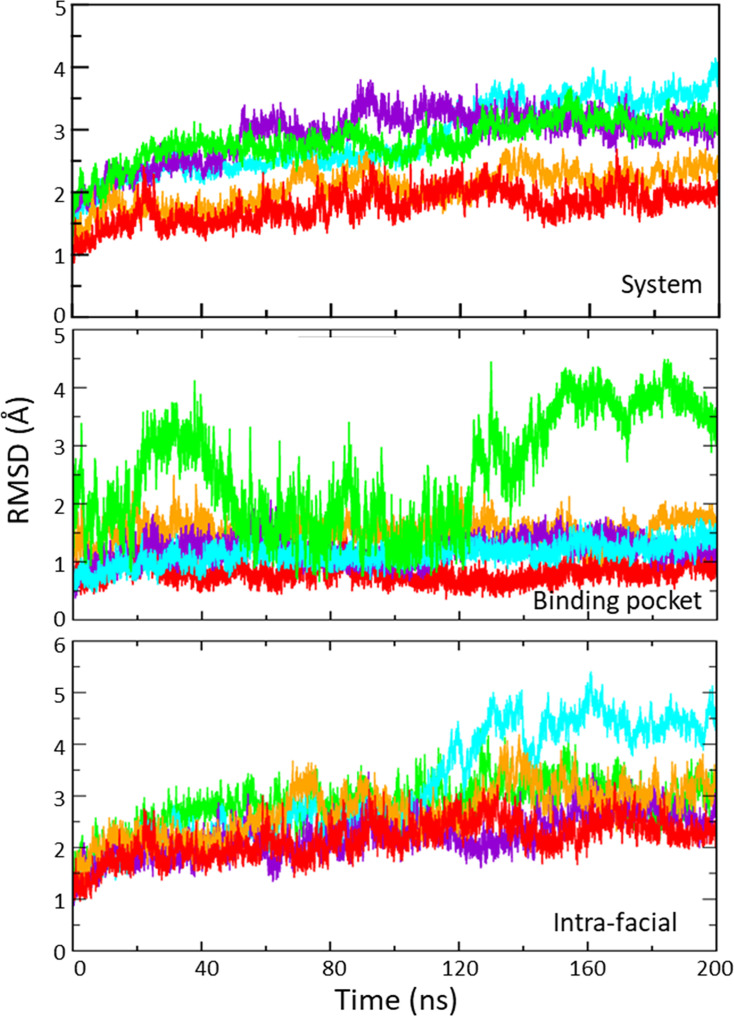
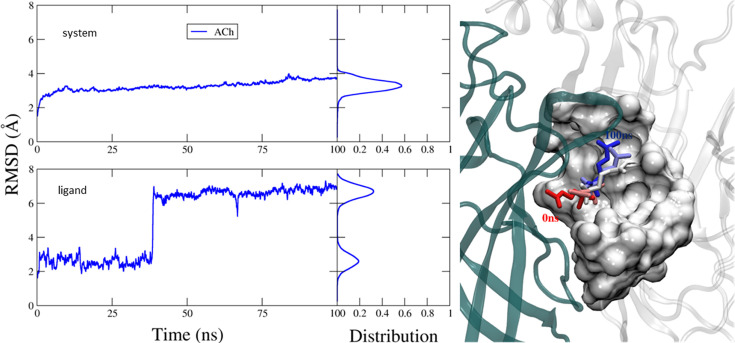
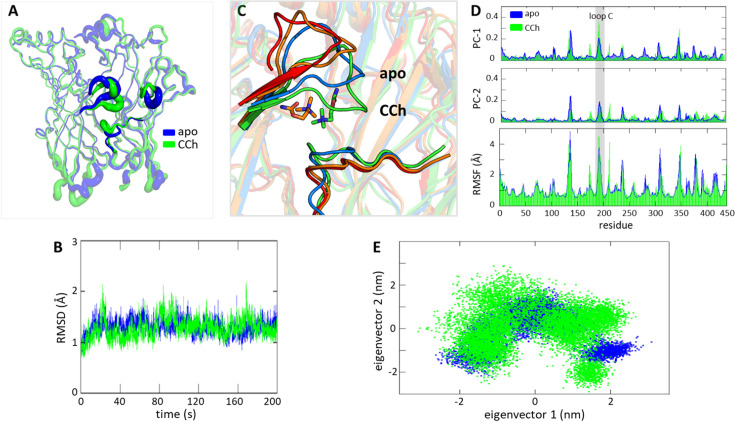
Figure 2. Agonist docking and loop dynamics.
(A) Top, agonists (blue, cationic center): carbamylcholine (CCh), acetylcholine (ACh), epibatidine (Ebt), and epiboxidine (Ebx). Bottom, α−δ site with docked agonists (top three poses). Resting-C, 6UVW.pdb minus toxin (red): loop C is up and agonist is cis; 200 ns, after simulation and removal of CCh (blue): loop C is down and agonist is trans. (B) Bottom, for all four agonists the docking scores (mean ± SD, n=3) were more favorable after simulation. (C) Cα root-mean-square deviation (RMSD) (mean ± SD, triplicates) are stable after ~120 ns (ACh, cyan; CCh, green; Ebt, orange; Ebx, purple). (D) Close-up of the CCh-occupied pocket. Red, resting-C; orange, equilibrated (0 ns molecular dynamics, MD); blue, after 200 ns MD. IN the simulations, loop C flops down (arrow), loop F moves in, and the agonist flips cis→trans (circled inset).