Extended Data Fig. 5 ∣. Stochasticity of T7 RNAP evolution.
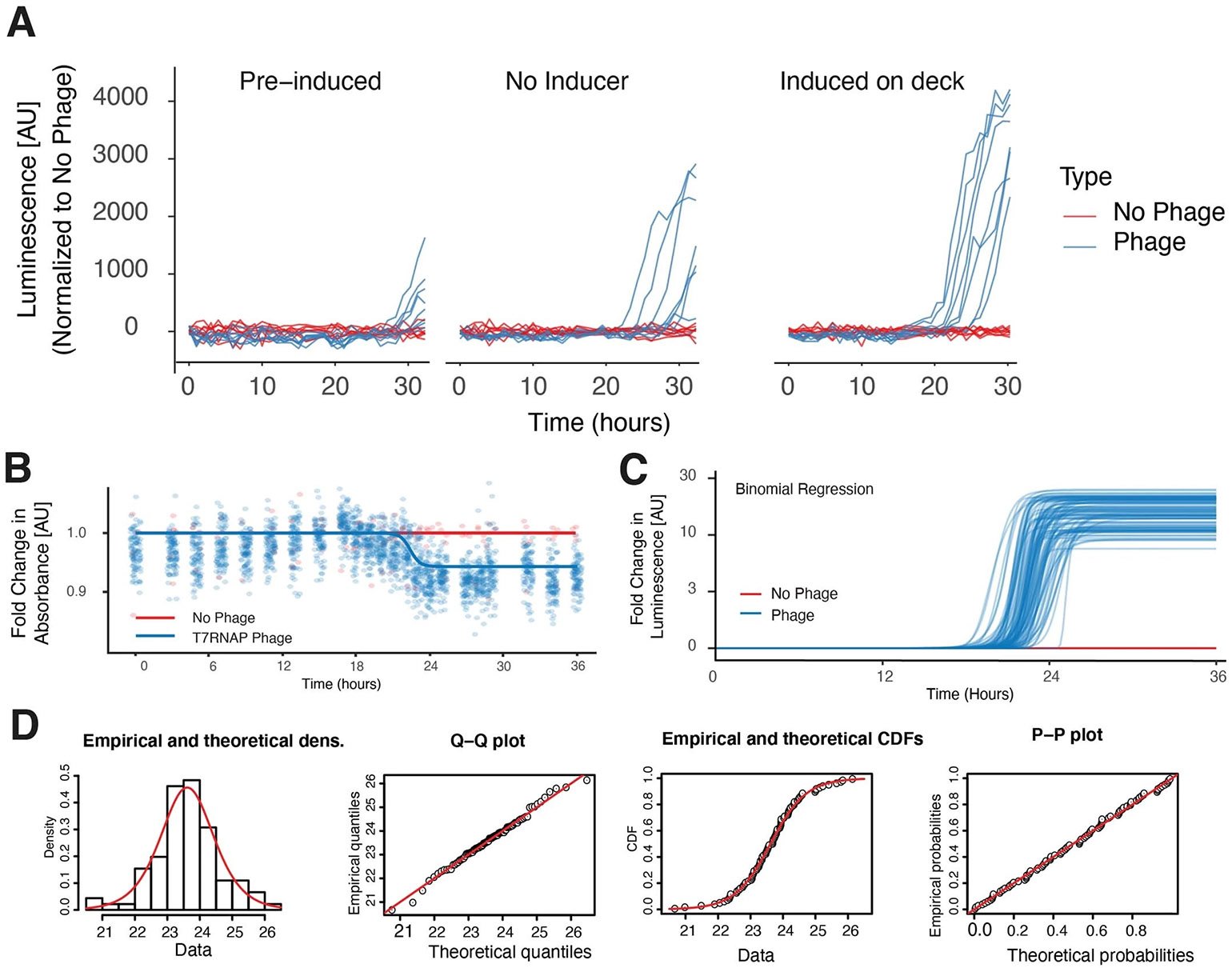
Validating T7 mutagenesis with cool PRANCE. MP-containing bacteria were provided with either 1) induction prior to cooling to 4 C, 2) given no inducer, or 3) induced on the robotic deck at 37 C and their luminescence was tracked for 30 hours to validate that mutagenesis behaved similarly to induction of cultures directly from a turbidostat (see Fig. 1d). (b) Real-time absorbance depression monitoring of 90 simultaneous directed evolution experiments with 6 no-phage controls, fit with a binomial regression of the total data. (c) Logistic regression of each luminescence trace during T7 RNAP evolution to bind the T3 promoter, used to calculate the average evolution times (Supplemental Methods). (d) Goodness-of-fit estimates of a logistic distribution of the total T7 evolution time data.
