Fig. 4 ∣. Feedback-controlled evolution of diverse starting genotypes.
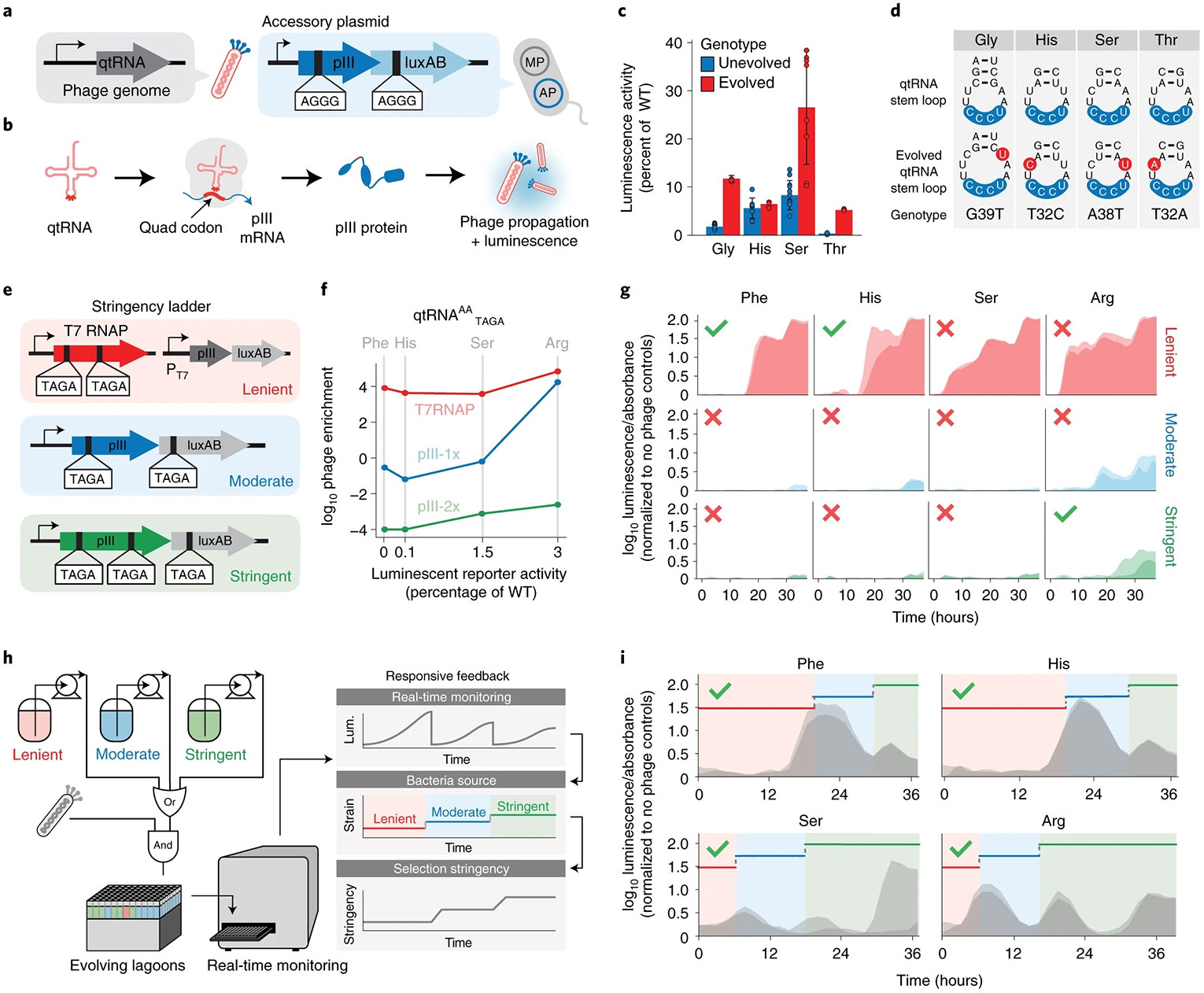
a, Strategy for evolving qtRNA to recognize quadruplet codons. AGGG quadruplet codons are inserted into the pIII protein and luxAB, and the evolving qtRNA are encoded in the evolving phage genome. AP, accessory plasmid. b, Phage propagation and luminescence are contingent on the successful decoding of the quadruplet codons in the pIII and luxAB proteins. Failure to decode a quadruplet codon results in premature termination and truncated protein. c, Efficiency of evolved versus unevolved qtRNA at incorporating amino acids compared to a triplet codon. d, Initial and evolved qtRNA genotypes. Data are presented as mean values ± s.e.m. for n = 3–8 biologically independent samples. e, Strategy for evolving qtRNAs with increasing stringency of selection (red/lenient, T7 RNAP; moderate/blue, pIII-1x; stringent/green, pIII-2x). f, The transfer function that determines the efficiency of phage propagation as a function of biomolecule activity for each AP, measured using phage-bearing qtRNAs at different starting activities (Phe, His, Ser, Arg). Starting activity is quantified as percentage of WT, or the luminescence generated by coexpressing the qtRNA and luxAB-357-TAGA, relative to an all-triplet luxAB. g, Evolution of four qtRNAs on APs with varying stringencies (successful evolution indicated by green checkmarks). h, In a feedback experiment, three bacterial strains with increasing stringency are added to phage populations and monitored in real time. The bacteria source for each well is adjusted in response to real-time luminescence measurements to automatically increase stringency of the environment. i, Real-time luminescence measurements of evolving qtRNAs with feedback-controlled selection stringency intervals.
