Figure 1: A neural network architecture to analyze structurally heterogeneous particles imaged by cryo-ET.
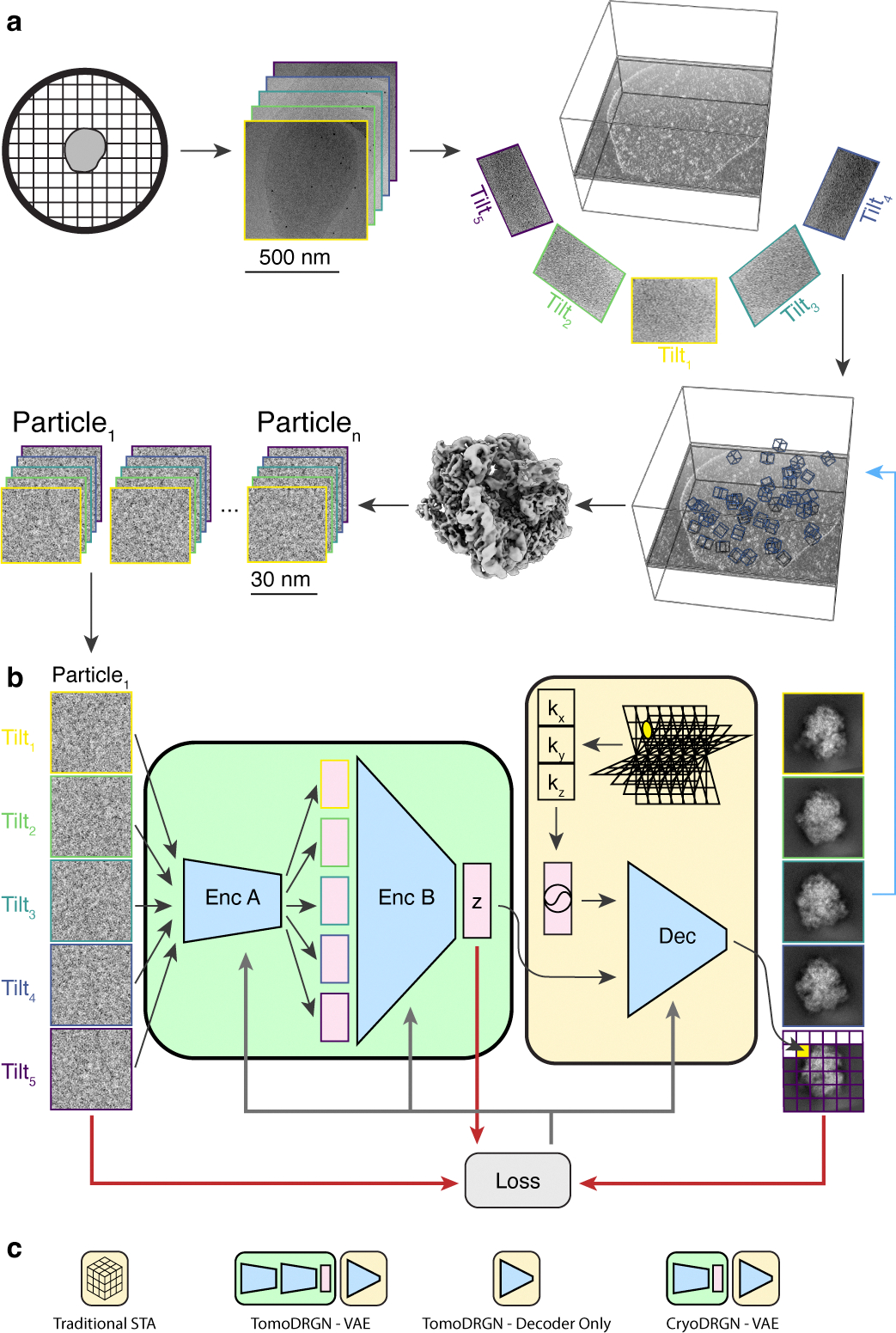
(a) A typical sample and data processing workflow to produce tomoDRGN inputs. The sample (e.g., a bacterial cell) is applied to a grid, plunge frozen, and optionally thinned. A series of TEM images of a target region are collected at different stage tilts. A tomographic volume is reconstructed using weighted back-projection of all tilt images. Instances of the target particle are identified (blue boxes) and extracted as 3-D voxel arrays. Iterative sub-tomogram averaging (STA) is used to reconstruct a consensus density map. Per-particle 2-D tilt images are then re-extracted from the source tilt series images and parameters (e.g. pose, defocus, etc.) estimated from STA are associated with the images.
(b) The tomoDRGN network architecture and training design. Each particle’s set of tilt images are independently passed through Encoder A, then jointly passed through Encoder B, thereby mapping all tilt images of a particle to one embedding (z) in a low dimensionality latent space. The decoder network (Dec) uses the latent embedding and a featurized voxel coordinate to decode a corresponding set of images pixel-by-pixel. Note that the decoder can learn a homogeneous structure by excluding the encoder module (green). The network is trained using a loss function (grey arrows) that depends on the input images, reconstructed images, and z (red arrows).
(c) Graphical signposts for volumes generated or analyzed by different reconstruction tools. These signposts are used throughout this manuscript when volumes are displayed to clarify how they were generated.
