Extended Data Fig. 4. TomoDRGN identifies non-ribosomal particles picked from EMPIAR-10499 tomograms.
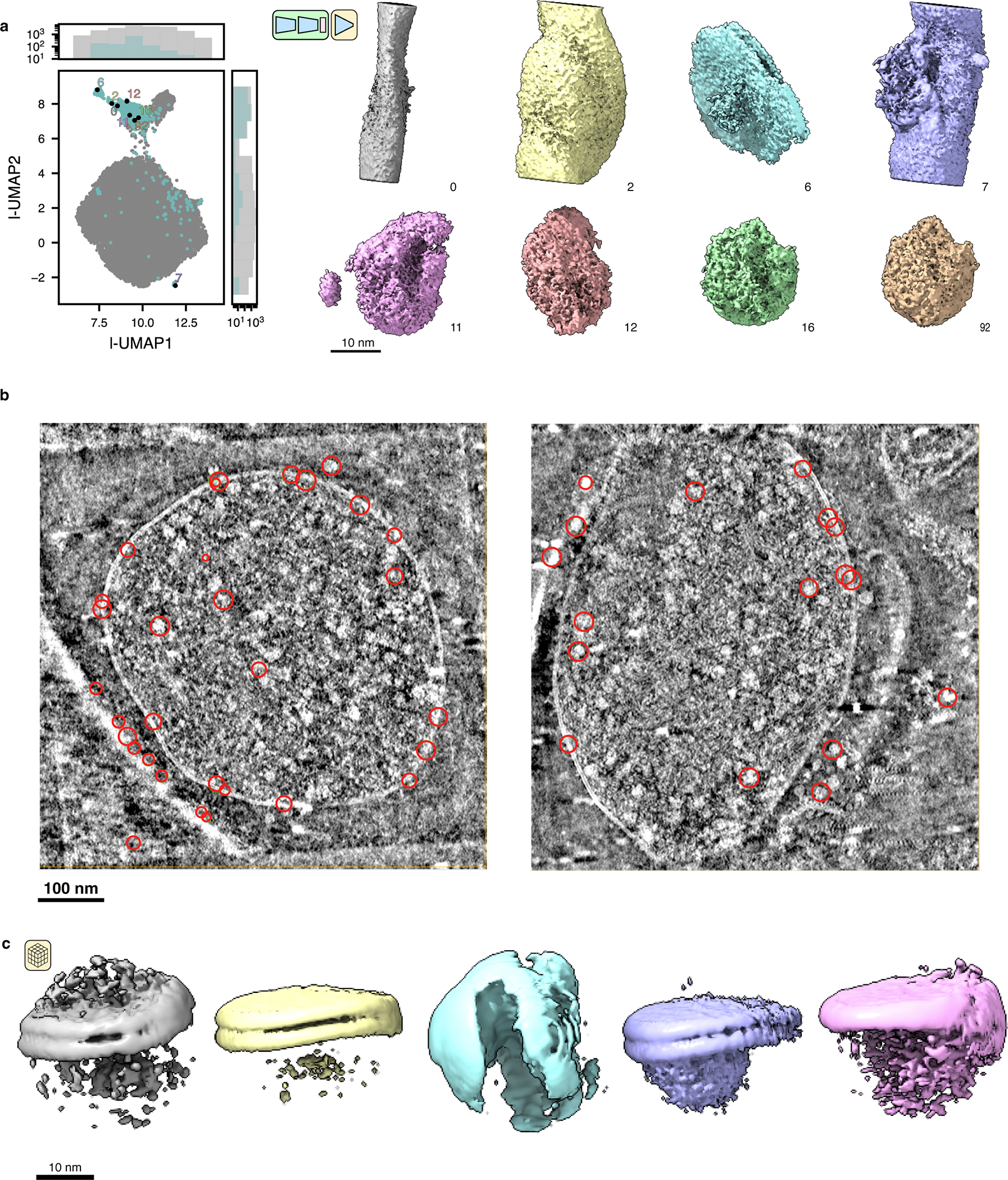
(a) Latent UMAP and corresponding sampled volumes from tomoDRGN heterogeneous network training from Fig. 5a. Eight representative non-ribosomal particles identified through manual inspection of k=100 k-means clustering of latent space are rendered at a constant isosurface and pose.
(b) Two tomograms are shown in slice view using Cube (https://github.com/dtegunov/cube) with locations of particles labeled as non-ribosomal annotated within each tomogram.
(c) RELION3-based multiclass (k=5) ab initio sub-tomogram volume generation using particles annotated as non-ribosomal via tomoDRGN (n=1,310).
