Extended Data Fig. 8. CryoDRGN learns errant structural heterogeneity in an exemplar tomographic dataset.
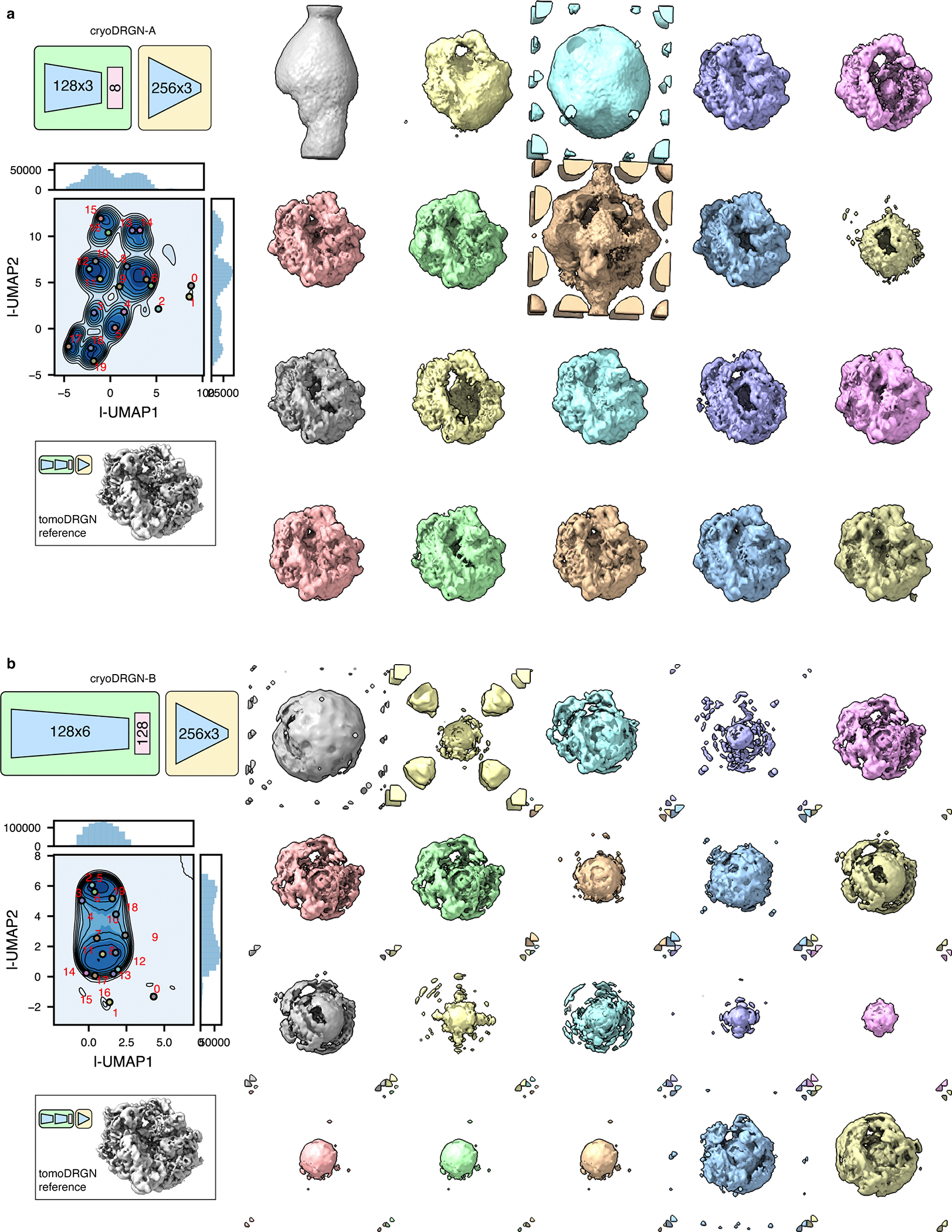
Two cryoDRGN models (a, b) were trained on the unfiltered particle stack of Mycoplasma pneumoniae ribosomes from Fig. 5a (n = 22,291 particles, treated as n = 913,931 images). The latent space is shown as a KDE plot following UMAP dimensionality reduction, with k=20 k-means class center particles annotated (left) and corresponding volumes visualized (right). Note that many putative 70S particles lack density in the particle core. A reference 70S volume sampled from tomoDRGN’s model in Fig. 5a is shown in the same pose for comparison.
