Extended Data Fig. 10. Assessment of tomoDRGN sensitivity to pose accuracy.
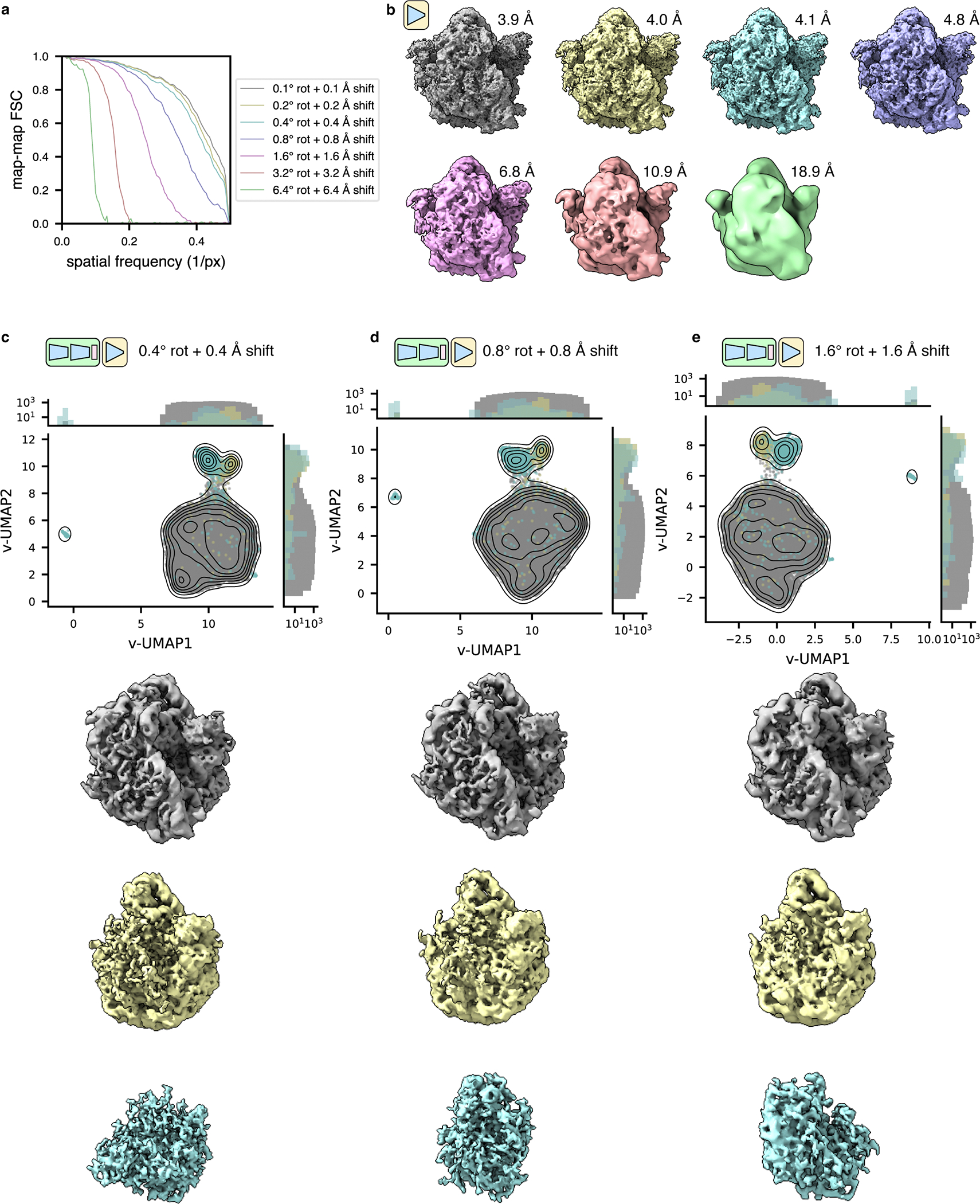
(a) The unfiltered stack of EMPIAR-10499 ribosomes in situ from Fig. 5a was used to train a series of tomoDRGN decoder-only models with increasing levels of random perturbations from STA-derived, “ground truth” rotation and translation poses (see Methods). The resulting map-map FSC curves against the STA ribosomal reconstruction are shown.
(b) Final tomoDRGN decoder-only reconstructed volumes corresponding to the FSC curves shown in (a). Volumes are lowpass filtered to the resolution where their map-map FSC to the STA ribosomal reconstruction crossed 0.5.
(c, d, e) UMAP of first 128 principal components of volume ensembles consisting of volumes generated for every particle, using tomoDRGN models trained on EMPIAR-10499 unfiltered ribosome stacks with indicated levels of pose perturbation. Particles annotated as 70S, 50S, and NR are colored as in Fig. 5c, with representative volumes of each class shown below. Note that NR particles are expected to be structurally diverse.
