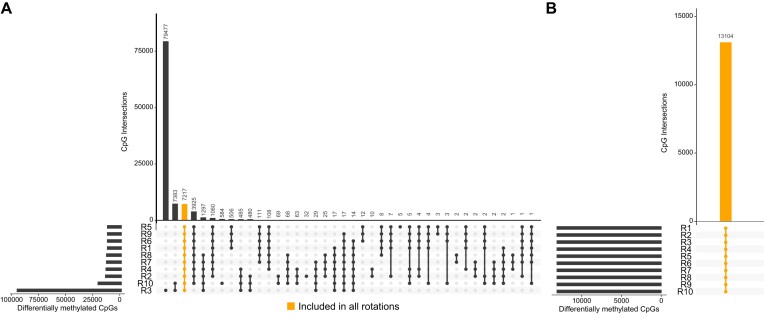
Figure 5.
Identification of differentially methylated CpGs while normalizing without and with a normalization sample. Six paired samples (n = 12), once cryo-preserved and once FFPE, were selected and randomly mixed to set up 10 different sample sheet rankings (R1–R10). Each ranking was once normalized without and with a normalization sample. Student t-test was applied to the 12 samples (cryo-preserved and FFPE) against 5 non-neoplastic samples for the 10 sample sheet rankings. (A) Intersections of differentially methylated CpGs (FDR < 0.01, mean |Δβ| > 0.3) normalized without a normalization sample. In addition to varying numbers of significant CpGs, only 7217 significant CpGs were detected in all sample sheet rankings. (B) Intersections of differentially methylated CpGs normalized with a normalization sample. The same 13 104 significant CpGs were detected for all sample sheet rankings.