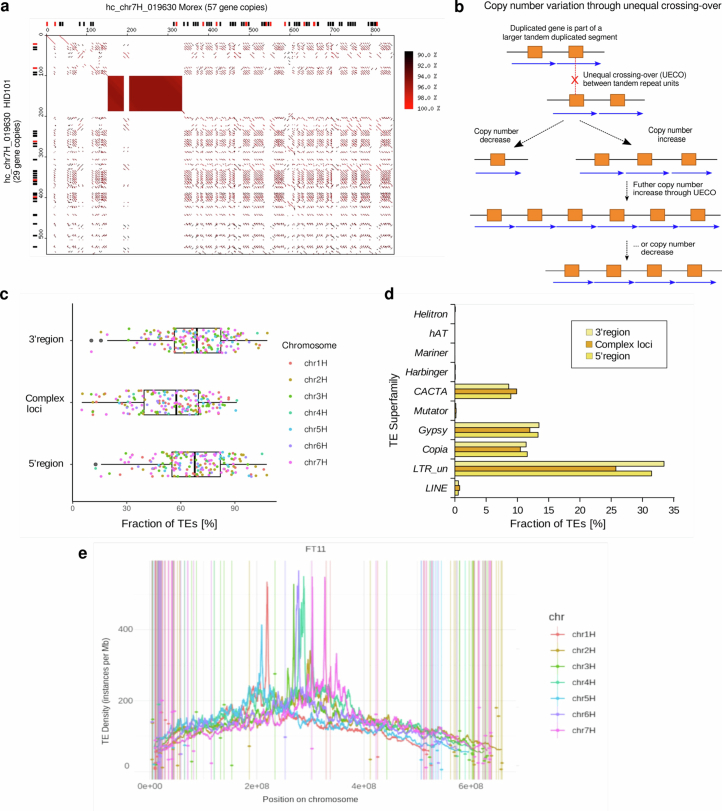
Extended Data Fig. 6. Complex loci are hot spots for copy number variation (CNV).
(a) Dot plot alignment of the example locus chr7H_019630 which contains a cluster of thionin genes. The sequences of Morex (horizontal) and wild barley HID101 (vertical) were aligned. Predicted intact genes are indicated as black boxes along the left and top axes. Predicted pseudogenes are shown in red. The axis scale is kb. The filled rectangle at positions ~150–330 kb in Morex represents an array of short tandem repeats which does not contain annotated thionin genes and does not have sequence homology to the thionin-containing tandem repeats of the locus. (b) The schematic model shows how, once an initial duplication is established, unequal homologous recombination (unequal crossing-over, UECO) between repeat units can lead to rapid expansion and contraction of the loci, thereby leading to CNV of genes. (c) TE content of complex loci. Dots represent the proportion of TEs (in %) in each to 169 complex loci. This is compared to regions of the same size (1 Mb) in the 3′ and 5′ directions. Complex loci have overall slighly lower content of annotated TEs than their flanking region, which is likely due to their higher gene content. Boxes indicate the inter-quartile range (IQR) with the central line indicating the median and whiskers indicating the minimum and maximum without outliers, respectively. Outliers were defined as minimum – 1.5 x IQR and maximum + 1.5 x IQR, respectively. (d) Contribution of TE superfamilies to complex loci and their 5′ and 3′ neigbouring regions. Complex loci contain slightly more CACTA and fewer LTR retroelements than neigbouring regions, a general characteristic of gene containing regions in barley. (e) Overall TE content along barley chromosomes (example accession B1K-04-12 [FT11]) compared to that of complex loci. TE content of complex loci is indicated by coloured dots. Due to the relatively small sizes of the loci, TE content of individual loci, in most cases, differs from that of the overall TE content in the respecive chromosomal regions.