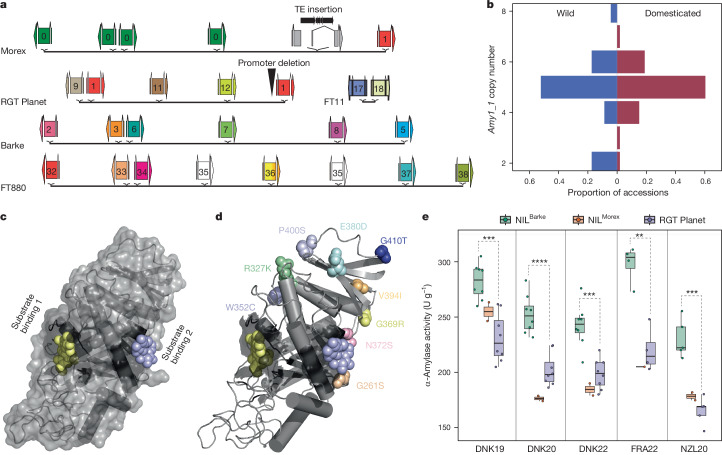
Fig. 3. Structural diversity at the amy1_1 locus and its importance in malting.
a, Simplified structure of the amy1_1 locus in selected pangenome assemblies. A detailed depiction of the amy1_1 locus across all 76 assemblies is shown in Extended Data Fig. 9a. Identical colours indicate identical ORFs in a and d. b, Distribution of amy1_1 copy numbers (as proportion of wild or domesticated accessions) across 76 assemblies. c,d, X-ray crystal structure (PDB 1BG9, ref. 39) of α-amylase bound to acarbose as a substrate analogue (magenta and yellow spheres). In d, amy1_1 amino acid variants (found in Morex, Barke and RGT Planet; Supplementary Table 21) are added as coloured spheres. e, α-Amylase activity of micro-malted grain of RGT Planet compared to RGT Planet near-isogenic lines (NILs) containing amy1_1-Morex and Barke haplotypes. The boxes delimit the 25th and 75th percentiles, and the horizontal line inside the box represents the median. Lower and upper whiskers denote minima and maxima. Two-sided t-test was used in multiple comparison and P value was adjusted with the Holm–Bonferroni method (**P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001). n = 8 (Barke), 2 (Morex), 8 (RGT Planet) independent samples examined in 5 independent experiments or environments.