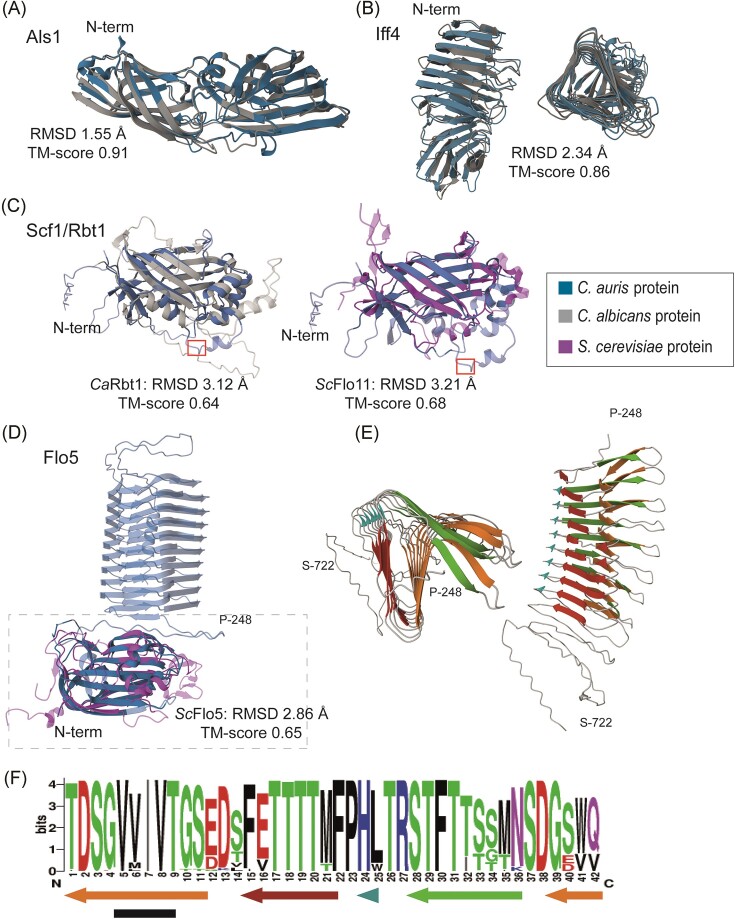
Figure 4.
Comparative structural analysis of C. auris adhesins. (A–D) Cartoon presentations of modeled three-dimensional structures (AlphaFold2) of putative ligand-binding domains of C. auris adhesins aligned (RCSB TM-alignment) with their closest C. albicans or S. cerevisiae homologs. (A) Als family protein Als1; (B) Iff/Hyr family protein Iff4, side and top views; and (C) Scf1/Rbt1. A canonical surface-exposed Kex2 cleavage site (KR/DV) at positions 216–219 in Scf1 is indicated by a red box. (D) Flo5 (strain B11220). The Flo5 model includes the repeat domain (top part) downstream of the ligand-binding domain (boxed), the latter aligned to ScFlo5. N-terminal residues in the structures of the C. auris proteins are indicated. (E) Top and side view of the strain B11220 Flo5 42-aa repeat domain (aa 248–722). (F) Sequence logo of the 42-aa repeat based on all Flo5 repeats in six representative strains (one per clade). Arrows indicate β-sheet forming regions, and the black bar marks a small region with high β-aggregation propensities according to TANGO. Ca, C. albicans and Sc, S. cerevisiae.