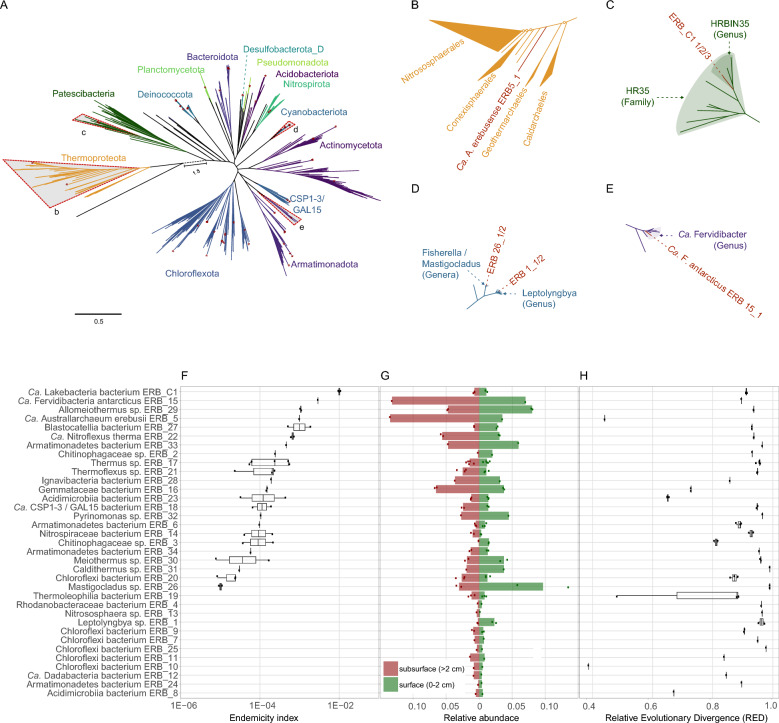
Fig. 3.
a Phylogenomic reconstruction of all MAGs recovered from Tramway Ridge, coloured according to phylum-level classification by GTDB-Tk v.2.4.0 using GTDB release 220. Trees were reconstructed with IQtree2 using an alignment of 34 concatenated marker genes generated with CheckM v.1.2.3. ModelFinder identified the best-fit model as LG + F + I + I + R10. Only MAGs with > 60% completeness and < 5% contamination as determined with CheckM v.1.2.3 were used for phylogenetic reconstruction. Bootstrap values have been omitted and location of Tramway MAGs are denoted with a red asterisk. Four clades of interest have been marked with grey triangles. The branch separating Archaea and Bacteria has been truncated and the actual branch length is given as 1.8. A complete, annotated tree is provided as Figure S1. b–e Subtrees from a that are bound by grey triangles. The majority of bipartitions shown exceed 95% UFboot support. Those marked with an open circle are those with less than 95% UFBoot support. Relevant taxonomic clades defined in GTDB release 220 are shaded and the location of Tramway MAGs are indicated as necessary for clarity. f Endemicity index (EI) of species-level collections of MAGs. Each line presents data for one species-level collection of MAGs (> 96.5% gANI). Strain-level MAGs within each species are shown as individual dots. Vertically, species are ordered by EI, with a higher EI indicating a larger diversity at synonymous coding positions. g Read abundance is reported as the relative proportion of metagenomic reads in Illumina datasets from the subsurface (brown–red) and near-surface (green) samples that map to each MAG. The middle line represents 0 and deviations to the left represent abundance in the subsurface and deviations to the right represent abundance in the near-surface. h Relative Evolutionary Divergence (RED) values for each MAG as determined with GTDB-Tk. Low RED values correspond to deep branches within the reference tree