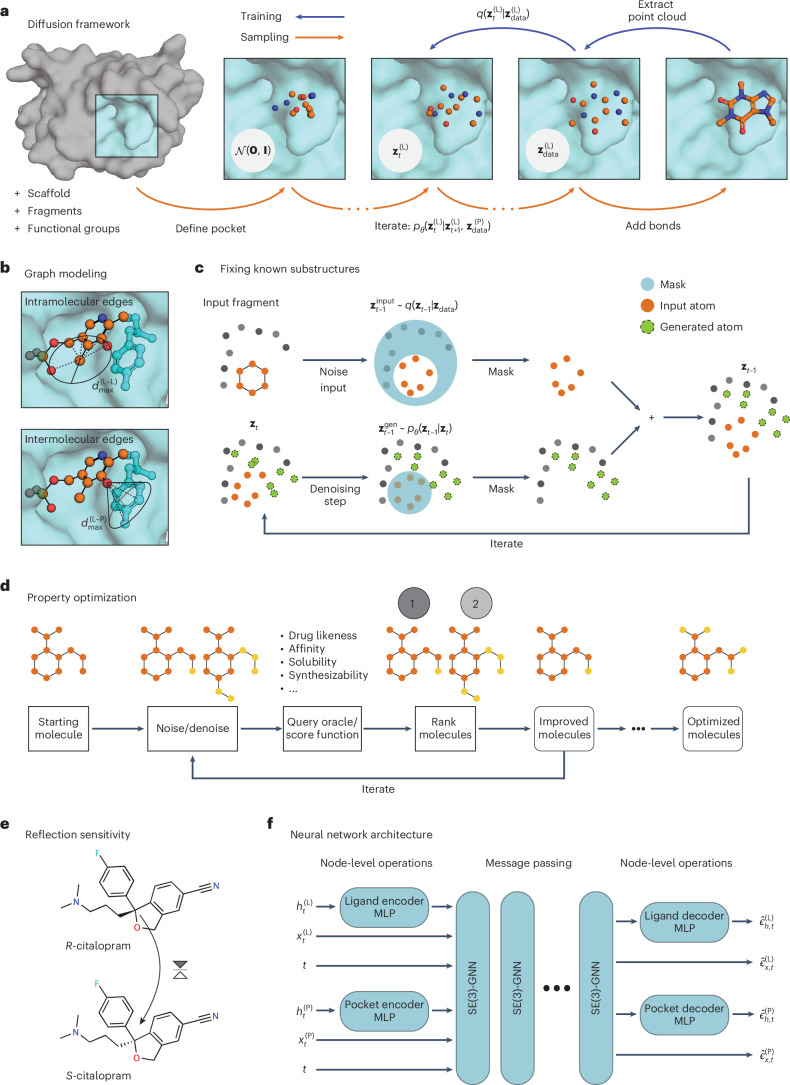
Fig. 1. Method overview.
a, The diffusion process q yields a noised version of the original atomic point cloud for a time step t ≤ T. The neural network model is trained to approximate the reverse process conditioned on the target protein structure . Once trained, an initial noisy point cloud is sampled from a Gaussian distribution and progressively denoised using the learned transition probability pθ. Covalent bonds are added to the resultant point cloud at the end of the generation. Optionally, fixed substructures (for instance, fragments) can be provided to condition the generative process. Carbon, oxygen and nitrogen atoms are shown in orange, red and blue, respectively. b, Each state is processed as a graph where edges are introduced according to edge type-specific distance thresholds, for instance, and . c, To generate new chemical matter conditioned on molecular substructures, we apply the learned denoising process to the entire molecule (superscript ‘gen’), but at every step we replace the prediction for the static substructure with the ground-truth noised version computed with q (superscript ‘input’). The protein context (gray) remains unchanged in every step. d, To tune molecular features, we find variations of a starting molecule by applying small amounts of noise and running an appropriate number of denoising steps. The new set of molecules is ranked by an oracle and the procedure is repeated for the best-scoring candidates. e, DiffSBDD is sensitive to reflections and can thus distinguish molecules with different stereochemistry. f, The neural network backbone is composed of MLPs that map scalar features h of ligand and pockets nodes into a joint embedding space, and SE(3)-equivariant message passing layers that operate on these features, each node’s coordinates x and a time step embedding t. It outputs the predicted noise values for every vertex.