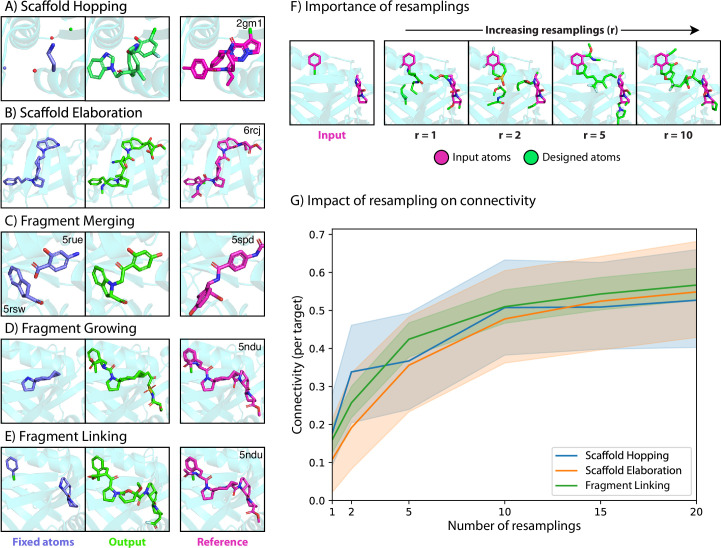
Extended Data Fig. 1. Molecular inpainting results.
Design examples for scaffold hopping (A), scaffold elaboration (B), fragment merging (C), fragment growing (D) and fragment linking (E). The inputs to our model (the fixed atoms) are shown in blue, the outputs (designed molecules) are shown in green and the original molecules are shown in magenta for reference. PDB codes are shown for the ground truth structure. In the case of fragment merging, we compose fragments with two different crystal structures with PDB codes shown. (F) Importance of resampling for generating realistic and connected molecules. The designed region (green) finally harmonizes with the molecular context at high resamplings. (G) Effect of the number of resampling steps on molecular connectivity. Carbon atoms are shown in light blue, green, or magenta depending on atom character. Oxygen, nitrogen, sulfur and chlorine are shown in red, dark blue, yellow, and light green, respectively. Means and 95% confidence intervals are plotted for 3 design tasks. For this experiment we used 20 randomly selected targets from the test set.