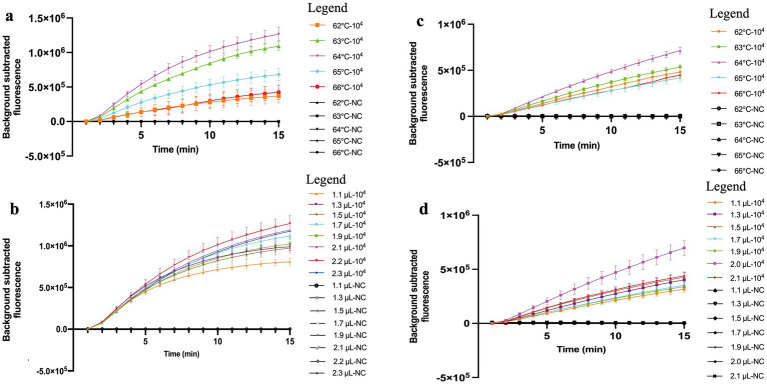
Figure 3.
Optimal reaction conditions of the COVID-19 MCTOP assay. Error bars represent the means ± standard error of means (SEM) from three replicates. (A) Optimal reaction temperature for ORF1ab gene detection. The reactions were performed under temperatures ranging from 62°C to 66°C at 1°C intervals. (B) Optimal primer concentration for ORF1ab gene detection. Serial volumes of MCDA primer premixture (2.2 μL and 1.1 to 2.3 μL with 0.2 μL intervals) were used to prepare the reaction mixtures. Primer concentrations for serial volumes were as follows. 1.1 μL (0.2 μM each of F1 and F2, 0.4 μM each of C1, C2, R1, R2, D1 and D2, 0.8 μM each of CP1 and CP2), 1.3 μL (0.24 μM each of F1 and F2, 0.47 μM each of C1, C2, R1, R2, D1 and D2, 0.95 μM each of CP1 and CP2), 1.5 μL (0.27 μM each of F1 and F2, 0.55 μM each of C1, C2, R1, R2, D1 and D2, 1.09 μM each of CP1 and CP2), 1.7 μL (0.31 μM each of F1 and F2, 0.62 μM each of C1, C2, R1, R2, D1 and D2, 1.24 μM each of CP1 and CP2), 1.9 μL (0.35 μM each of F1 and F2, 0.69 μM each of C1, C2, R1, R2, D1 and D2, 1.38 μM each of CP1 and CP2), 2.1 μL (0.38 μM each of F1 and F2, 0.76 μM each of C1, C2, R1, R2, D1 and D2, 1.53 μM each of CP1 and CP2), 2.2 μL (0.4 μM each of F1 and F2, 0.8 μM each of C1, C2, R1, R2, D1 and D2, 1.6 μM each of CP1 and CP2), 2.3 μL (0.42 μM each of F1 and F2, 0.84 μM each of C1, C2, R1, R2, D1 and D2, 1.67 μM each of CP1 and CP2). (C) Optimal reaction temperature for N gene detection. The reactions were performed under temperatures ranging from 62°C to 66°C at 1°C intervals. (D) Optimal primer concentration for N gene detection. Serial volumes of MCDA primer premixture (2.0 μL and 1.1 to 2.1 μL with 0.2 μL intervals) were used to prepare the reaction mixtures. Primer concentrations for serial volumes were as follows. 1.1 μL (0.22 μM each of F1 and F2, 0.44 μM each of C1, C2, R1, R2, D1 and D2, 0.88 μM each of CP1 and CP2), 1.3 μL (0.26 μM each of F1 and F2, 0.52 μM each of C1, C2, R1, R2, D1 and D2, 1.04 μM each of CP1 and CP2), 1.5 μL (0.3 μM each of F1 and F2, 0.6 μM each of C1, C2, R1, R2, D1 and D2, 1.2 μM each of CP1 and CP2), 1.7 μL (0.34 μM each of F1 and F2, 0.68 μM each of C1, C2, R1, R2, D1 and D2, 1.36 μM each of CP1 and CP2), 1.9 μL (0.38 μM each of F1 and F2, 0.76 μM each of C1, C2, R1, R2, D1 and D2, 1.52 μM each of CP1 and CP2), 2.0 μL (0.4 μM each of F1 and F2, 0.8 μM each of C1, C2, R1, R2, D1 and D2, 1.6 μM each of CP1 and CP2), 2.1 μL (0.42 μM each of F1 and F2, 0.84 μM each of C1, C2, R1, R2, D1 and D2, 1.68 μM each of CP1 and CP2). Ultrapure water served as negative control (NC).