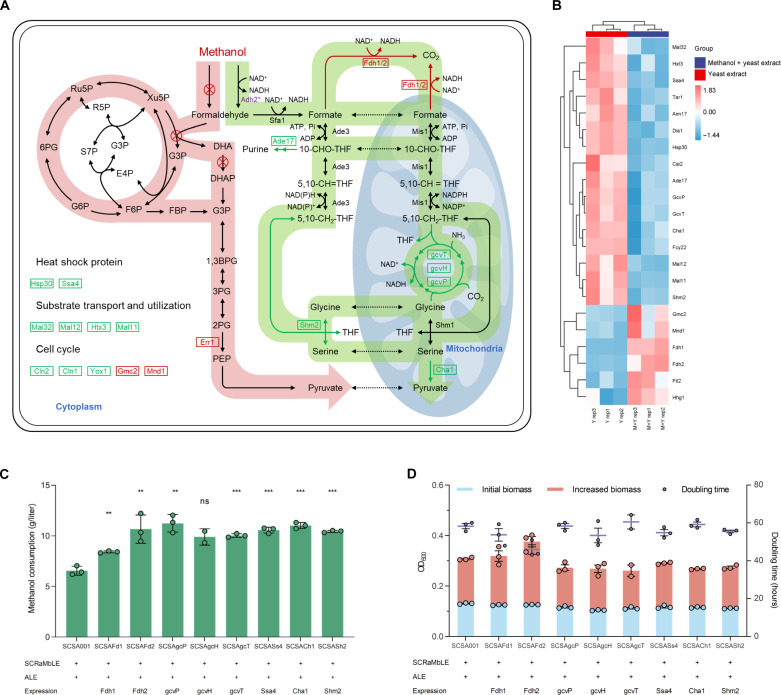
Fig. 4. Transcriptomic analysis and the validation of reverse engineering of the rGly pathway.
(A) Differentially expressed enzymes related to central metabolism and their effect on methanol utilization. The pathway with red background represents the heterologous methanol assimilation pathway, which was eradicated in SCSA001 and did not exhibit significant differences in expression. The pathway with green background represents the possible methanol utilization pathway in SCSA001, in which Fdh1/2 is significantly up-regulated and the rGly pathway is significantly down-regulated. Pi, inorganic phosphate; 10-CHO-THF, 10-Formyltetrahydrofolate. (B) Cluster analysis related to differentially expressed enzymes. Yeast cells for transcriptional analysis were cultivated and collected in Delft MM with 0.1% yeast extract or 0.1% yeast extract and 2% methanol for 48 hours. (C and D) Methanol consumption, biomass (OD600) formation, and doubling time after 72 hours of cultivation in Delft MM with 2% methanol (growth curves are shown in fig. S20). All strains were precultured in YPD medium and washed twice using PBS (pH 7.2) before cultivated in Delft MM. All data represent the mean ± SD. Triplicate independent samples were adopted for each panel. Statistical analysis was performed using a two-tailed Student’s t test (**P < 0.01, and ***P < 0.001). ADP, adenosine diphosphate.