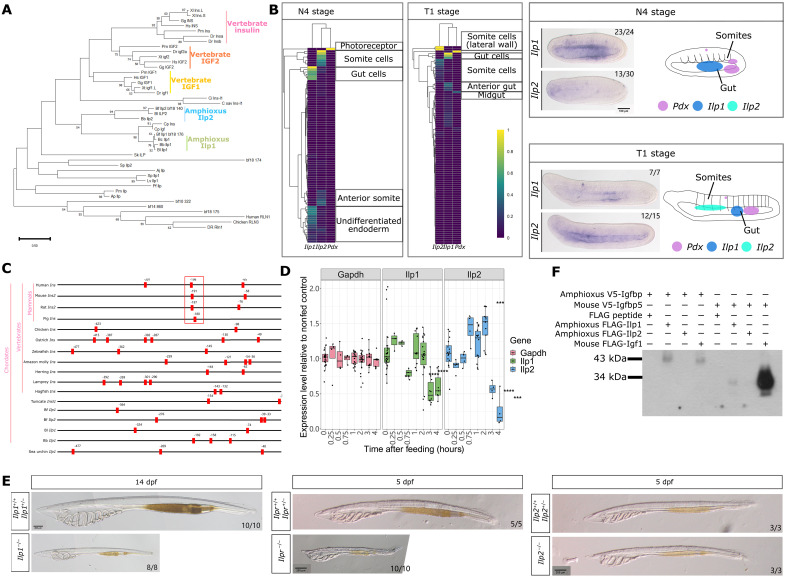
Fig. 5. Amphioxus Ilp1, Ilp2, and Ilpr function.
(A) Phylogenetic relationship of vertebrate insulin, Igf, and invertebrate Ilp constructed using maximum likelihood analysis. Vertebrate relaxin is used as an outgroup. (B) Expression of Ilp1 and Ilp2 in mid-neurula (N4 stage) and late neurula (T1 stage) amphioxus embryos. Normalized expression at the single-cell level is summarized in heatmaps, with Pdx expression shown for reference. In situ hybridization results for Ilp1 show expression in the gut endoderm in N4 embryos, which becomes localized to a small region in the gut in T1 embryos. Ilp2 expression is not detected in N4 embryos whereas expression in the anterior pharyngeal region is detected in T1 embryos. A rough summary of Pdx, Ilp1, and Ilp2 expression in N4 and T1 stage embryos, with Pdx expression patterns obtained from published in situ results (43, 67), is provided with areas of gene expression indicated using different colors. Scale bar, 100 μm. (C) Location of the Pdx-core binding motif TAAT in the 500-bp upstream region of vertebrate insulin genes and invertebrate Ilp genes. The highly conserved Pdx-binding A3 box region in mammals is highlighted using a red box. (D) Relative expression level of Gapdh, Ilp1, and Ilp2 genes in starved wild-type larvae stimulated with food compared to the seawater control. Statistical analysis was performed using two-sided t tests; **P = between 0.01 and 0.001, ***P = between 0.001 and 0.0001, ****P = below 0.0001. (E) Phenotype of Ilp1 mutant larvae 14 days after fertilization. Scale bar, 200 μm. (F) Interaction between amphioxus Ilp1, Ilp2, Igfbp, and mouse Igf1 and Igfbp5 proteins.