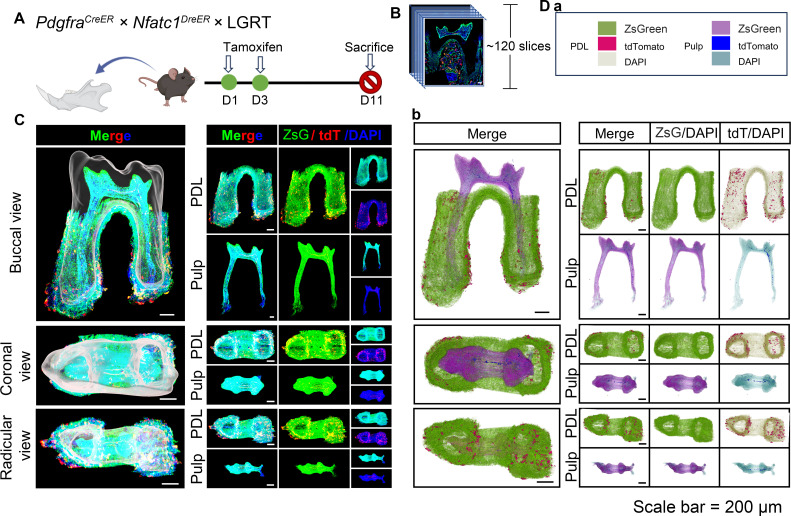
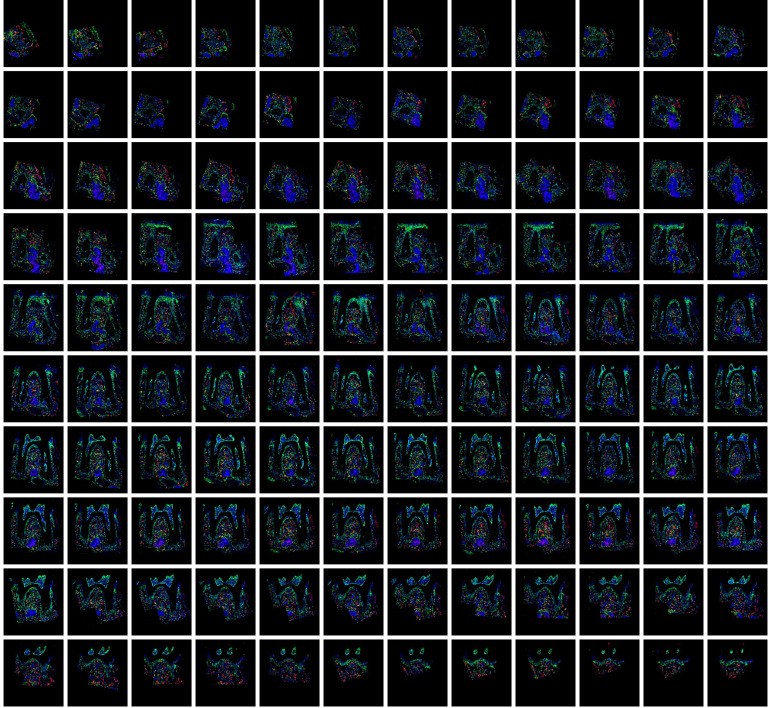
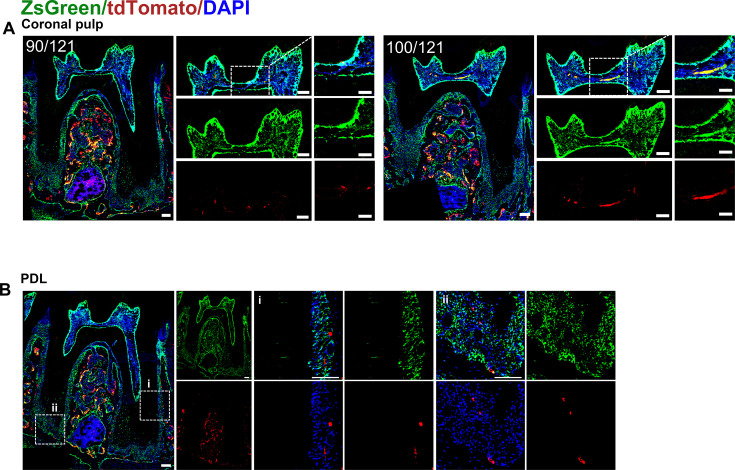
Figure 7. MCIR tracing distinct cell populations simultaneously in the pulp and PDL.
(A) Schematic illustration of pulse in PdgfraCreER×Nfatc1DreER× LGRT mice. (B) Operation process of frozen section. Using this procedure, a total of 120 slices were collected in this sample. (C) 3D reconstruction of mandible M1 using Imaris, including pulp and PDL with virtual dentin shell (white) (scale bar = 200 μm). The image stack was displayed in buccal view, coronal view, and radicular view of pulp and PDL, respectively. (D) 3D reconstruction of mandible M1 using DICOM-3D, in which the distribution of NFATc1+ cells in pulp and PDL tissues can be more precisely perceived (D-b), scale bar = 200 μm. (D-a): The legend of (D-b).