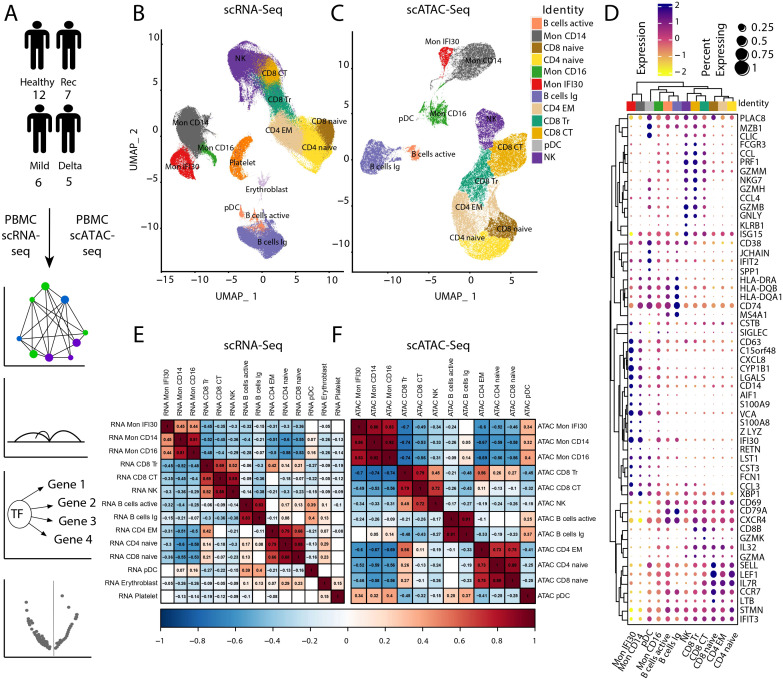
Figure 1.
Data analysis workflow and scRNA-seq/scATAC-seq integration. (A) We performed scATAC-seq of the PBMC samples from 12 healthy, seven convalescence, six mild COVID-19, and five individuals with severe/critical Delta COVID-19, followed by computational analysis and integration with scRNA-seq, reconstruction of the gene regulatory networks, and analysis of differential chromatin accessibility. (B) UMAP of scRNA-seq and cell-type annotation. (C) UMAP of scATAC-seq data and cell type annotation. (D) Signal of the Chromatin accessibility signal for PBMC marker genes. (E) Pseudobulk gene expression across different cell types using scRNA-seq. The gaps are correlation values with low significance level (p-value ). (F) Pseudobulk-approximated gene activity Pearson’s correlation across cell types in scATAC-seq. Gaps are correlation values with low significance level (p-value ).