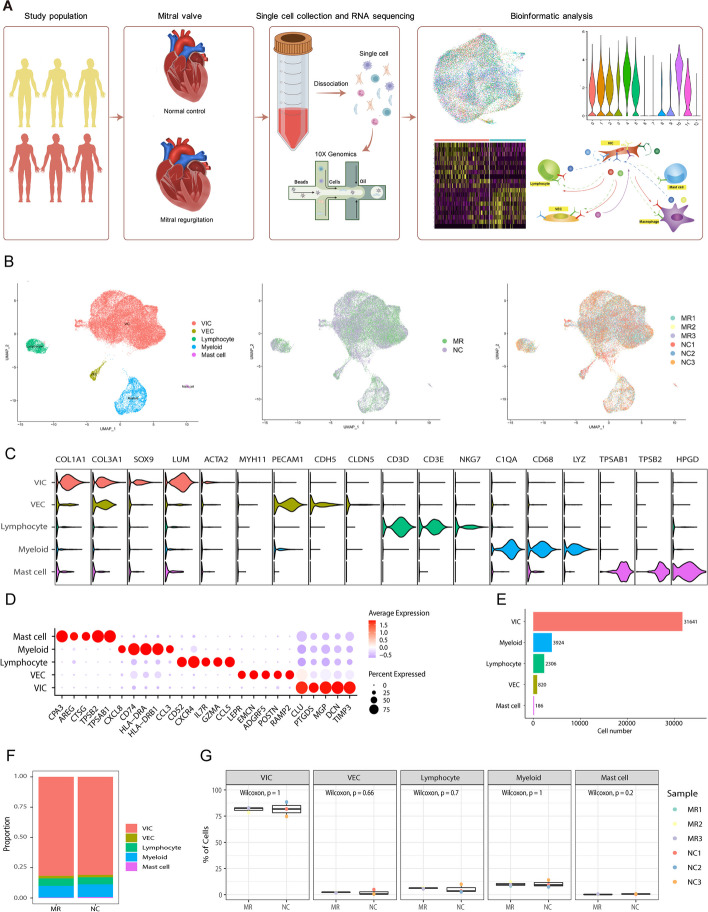
Fig. 1.
Clustering and identification of cell types in nondiseased and functional regurgitated human mitral valves with scRNA-seq data. A Six mitral valves from three normal controls and three patients with MR were harvested, and scRNA-seq was separately performed. B The UMAP plot of the combined six samples shows clusters, sample types, individual patients, and cell types. C The gene markers and each cell type identified are shown in the violin plot. D Dot plot of the top 5 marker genes in VIC clusters. The dot size corresponds to the proportion of cells within the group expressing each gene, and the dot color corresponds to the expression level. E The number of cells measured in six specimens. F The composition of each cell type in the NC and MR groups. G Box plots comparing the proportions of each cell type between the two groups. MR, mitral regurgitation; scRNA-seq, single-cell RNA sequencing; UMAP, Uniform Manifold Approximation and Projection; VIC, valvular interstitial cell; VEC, valvular endothelial cell