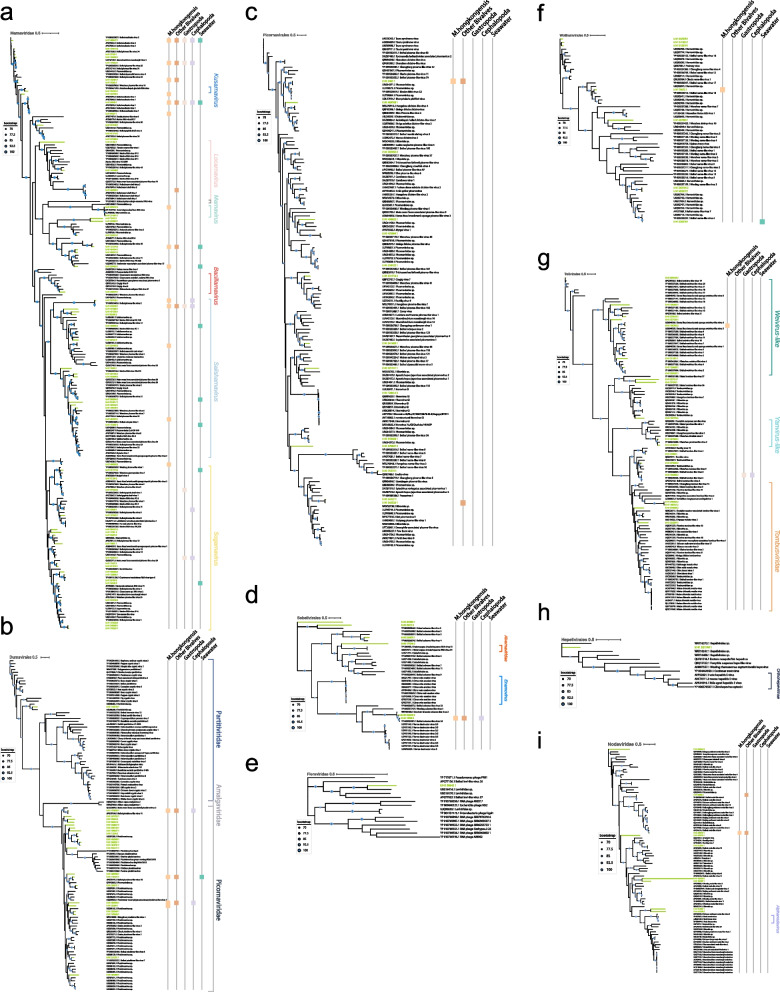
Fig. 2.
Phylogenetic analysis of the viruses (RdRPs) from M. gigas DT and related viruses from the NCBI nonredundant (nr) database. Unrooted phylogenetic trees are constructed using maximum-likelihood estimation and 1000 iterative refinements. Only bootstrap values greater than 0.7 are displayed in the tree. Green lines indicate the RNA viruses identified in this study. The scale bar indicates the average number of amino acid substitutions per site. If a specific virus is identified as the dominant virus in other meta-transcriptomes, it will be represented by a differently colored square on the right. a, b, c, d, e, f, g, h, i The phylogenetic tree of the following viral lineages: Marnaviridae, Durnavirales, Picornavirales, Sobelivirales, Fiersviridae, Wolframvirales, Tolivirales, Hepelivirales, and Nodaviridae, respectively