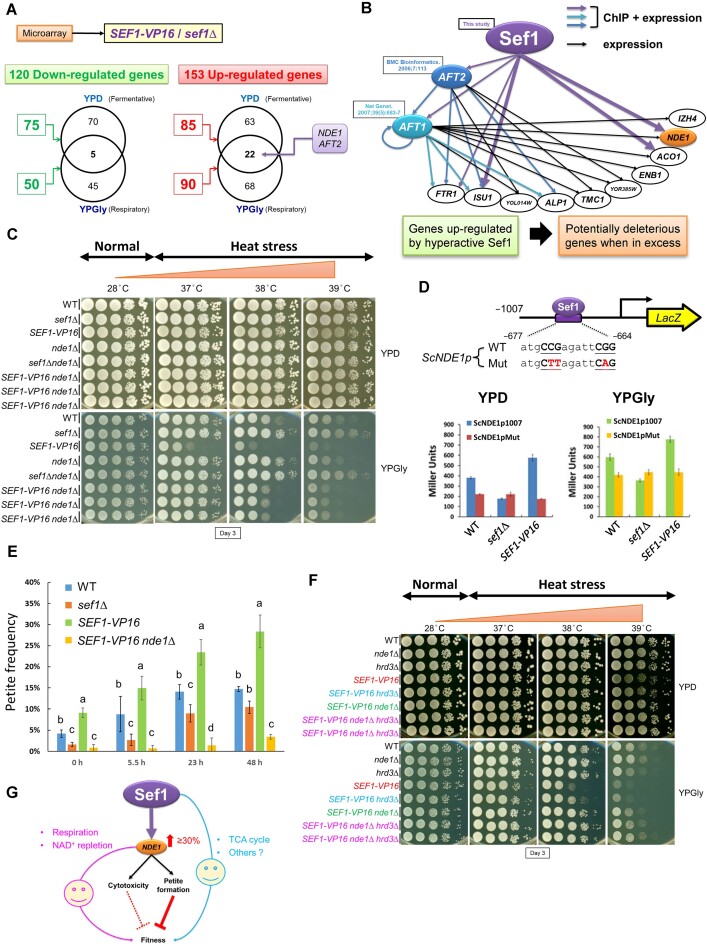
Figure 3.
NDE1 is the phenotype-determining gene of the Sef1-mediated transcription regulatory network in S. cerevisiae. (A) Summarized total numbers of differentially expressed genes in response to the hyperactive SEF1-VP16 versus the sef1Δ mutant under YPD and YPGly conditions. There are 120 downregulated and 153 up-egulated genes under either condition. Notably, NDE1 and AFT2 are upregulated under both conditions. (B) The Sef1-Aft2 subnetwork is composed of three TFs (including Sef1, Aft2 and Aft1) and ten SEF1-VP16 upregulated genes (Supplementary Tables S4–S7), which are also either Aft2 or Aft1 target genes and have been reported as deleterious at higher abundance (from Saccharomyces Genome Database phenotypes). The edges (expression and ChIP-binding connections) of this network are built according to data from this study (Supplementary Table S1) and previous reports (85,86). NDE1 is highlighted in orange. (C) Growth of SEF1-VP16 nde1Δ mutants in response to YPD, YPGly and heat stress in S. cerevisiae. All plates were incubated for 3 days. Three independent SEF1-VP16 nde1Δ clones were tested. Deletion of NDE1 can considerably rescue the growth defects generated by hyperactive Sef1. (D) Design of the verification system for Sef1 binding on the NDE1 promoter by the LacZ reporter-based promoter assay. Two consensus Gs and one consensus C were mutated to A or T (highlighted in red) (upper panel). The ChIP peak regions for the S. cerevisiae NDE1 promoters (−1 to −1007 from ATG) contain only one high-confidence (P-value <0.0001) Sef1 binding motif (Supplementary Figure S1D). The wild-type and motif-mutated promoters were fused to the LacZ reporter gene. The contribution of Sef1 motifs to promoter activities was assayed in wild-type, sef1Δ and hyperactive SEF1-V16 cells (bottom panel). LacZ activity was measured under both YPD and YPGly conditions using a liquid-galactosidase assay. The β-galactosidase levels are displayed as average Miller units ± SD from three technical repeats. Compared to the wild-type strain with the wild-type NDE1 promoter, Sef1 binding contributes to ∼40% and ∼30% of promoter activities under YPD and YPGly conditions, respectively. Moreover, the enhanced Sef1 activity from VP16 contributes to ∼50% and ∼30% of additional promoter activities under YPD and YPGly conditions, respectively. (E) The petite-inducing effects of Sef1 and Nde1. Petite formation was induced by growth in YPD at 39°C. The petite cells were counted at the indicated time points, and petite frequency is displayed as mean ± SD from five technical repeats. One-way ANOVA followed by Tukey’s multiple comparisons post hoc test was performed, with letters indicating significant differences. Petite formation rates increased with increasing Sef1 activity (sef1Δ, wild type, to SEF1-VP16) and required the presence of Nde1 (SEF1-VP16 nde1Δ). (F) Growth of the SEF1-VP16 hrd3Δ mutants in response to YPD, YPGly and heat stress in S. cerevisiae. All plates were incubated for 3 days. Two independent SEF1-VP16 nde1Δhrd3Δ clones were tested. Abolishing the Hrd3 function enhances the stability of mitochondrial DNA (mtDNA), thereby reducing the petite formation rate (45). Deletion of HRD3 can partially rescue the defective growth caused by Sef1 hyperactivity, despite this rescue effect being weaker than the effect of nde1Δ. There is no clear additive rescue effect when both HRD3 and NDE1 are deleted. (G) A working model depicting that ScSef1-mediated NDE1 expression determines cellular fitness via multiple moonlighting mechanisms, including two beneficial functions in respiration and NAD+ repletion and two deleterious functions in cytotoxicity (only when ultra-highly expressed) (44) and petite formation. Modulating the activities of Sef1 generally affects fitness by changing Nde1-driven petite formation rates in the population. In the presence of Nde1, the potentially positive effects of the other Sef1-regulated genes on fitness are much less significant.