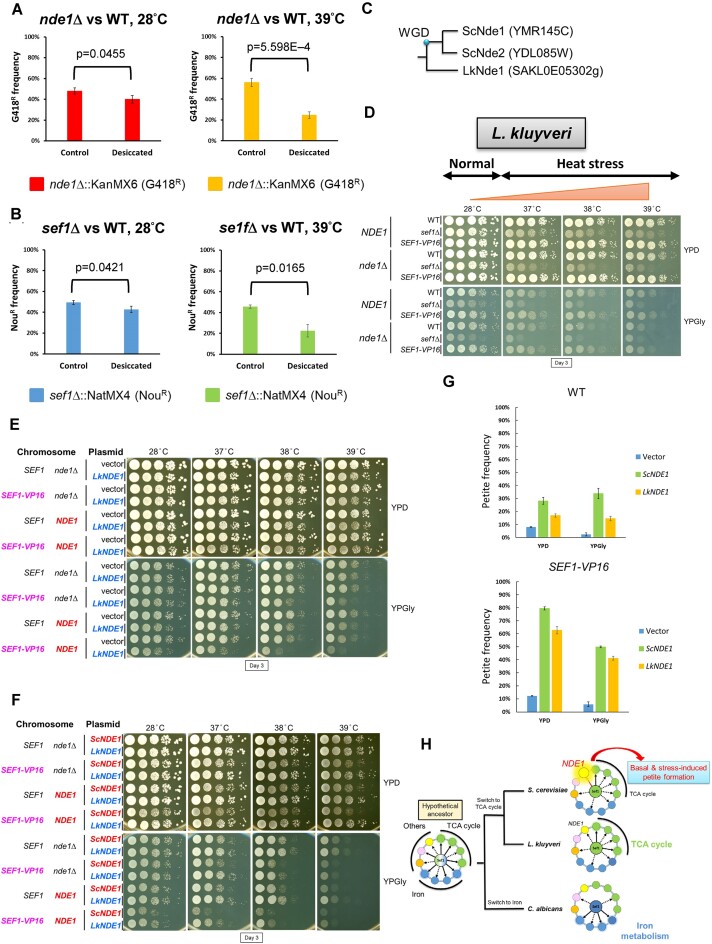
Figure 4.
Saccharomyces cerevisiae is more vulnerable to the petite-inducing effect of Nde1. (A) Nde1 contributes to desiccation tolerance. (B) Sef1 contributes to desiccation tolerance. For (A) and (B), desiccation assays were performed by mixing the drug-resistant deletion strains with the reference drug-sensitive wild-type strain at an initial 50:50 cell number ratio and air-dried for 24 h at 28 or 39°C. The frequency of the drug-resistant cells is shown. A two-tailed Student’s t-test was used to calculate the P-value for each pair by comparing the drug-resistant frequency of the control sample (without desiccation) with the desiccated sample. (C) Orthology of Nde1 between S. cerevisiae and L. kluyveri. There is only one Nde1 homolog in L. kluyveri. LkSef1 also binds to the NDE1 promoter (23). (D) Growth of the SEF1-VP16 nde1Δ mutants in response to YPD, YPGly and heat stress in L. kluyveri. All plates were incubated for 3 days. Deletion of NDE1 has no clear effect on cell growth, even under respiratory conditions. (E) Ectopic expression of the LkNDE1 homolog on a centromeric plasmid selected by HGB in S. cerevisiae. Relative to vector-only controls, additional expression of LkNDE1 exacerbates the growth defect, especially in strains harboring SEF1-VP16 and/or a chromosome copy of ScNDE1. (F) Ectopic LkNDE1 is less deleterious than ScNDE1 when additionally expressed in S. cerevisiae. For (E) and (F), cells were spotted and grown in YPD or YPGly, and with/without heat stress in the presence of 300 μg/ml HGB. All plates were incubated for 3 days. (G) Basal petite-inducing effects in response to ectopic expression of LkNDE1 and ScNDE1. Petite formation was induced by growth in YPD + HGB at 28°C for 20 h. Petite cells were counted by spreading them on YPD plates. Petite frequency is displayed as mean ± SD from five technical repeats. Relative to ectopic ScNDE1, ectopic LkNDE1 is less petite-inducing in both the wild-type and SEF1-VP16 backgrounds. (H) Proposed switches in Sef1 function from a hypothetical common ancestor to the non-regulating branch (C. albicans) and TCA cycle-regulating branch (23). The divergence between L. kluyveri and S. cerevisiae leads to a split of the true-TCA cycle regulating sub-branch and the pseudo-TCA cycle regulating sub-branch, with the phenotypic outcomes of this latter being dominated by the petite-inducing activity of the conserved Sef1 target gene, NDE1.