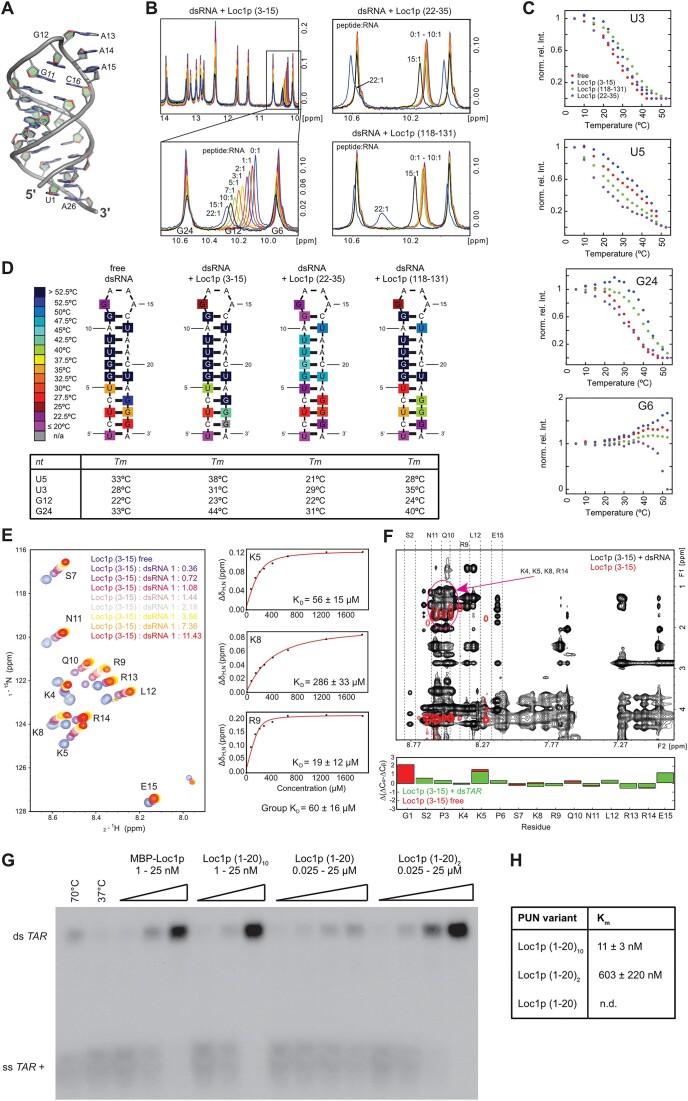
Figure 4.
Structure analysis of RNA binding and assessment of annealing activity of PUN motifs. (A) X-ray structure of the dsRNA tetraloop highlighting the coordinated GAAA minimal tetraloop (G12–A15) and the base-paired stem. (B) Titration of the dsRNA with different Loc1p-derived peptides. Whereas the titration of Loc1p (3–15) shifts the signal of G12 in a continuous manner, the two peptides lacking the RNA-binding motif Loc1p (118–131) and Loc1p (22–35) do not bind until very high, 15-fold excess, where peaks start to display sudden large shifts. (C) Temperature dependence of imino NMR signals of unbound dsRNA and dsRNA bound to different Loc1p-derived peptides at the highest molar excess (22:1) shown in panel (B). The decrease of imino signal intensities (shown: U3, U5, G6, G24) was used to determine the stabilizing effect of the three different peptides titrated in panel (B) in temperature scans. Whereas Loc1p (3–15) (blue dots) has a stabilizing effect on the hairpin RNA (free RNA: red dots), the control peptides Loc1p (118–131) (green dots) and Loc1p (22–35) (purple dots) have an overall destabilizing effect. The only exception is a slight stabilizing effect for U3 by Loc1p (118–131). (D) Heat map illustrating the temperature-dependent stabilization of the end of the stem upon peptide binding. In contrast, Loc1p (118–131) and especially Loc1p (22–35) destabilize the ends of the stem and the loop regions. Loc1p (22–35) has even a destabilizing effect on the central stem region. The estimated melting temperatures for the four most affected base pairs are given in the table below. (E) 1H,15N-HSQC spectrum showing the titration of dsRNA to 15N,13C-labeled Loc1p peptide. Saturation is reached at a 3–4-fold excess of dsRNA tetraloop. To the right, three examples are shown of fitted binding curves from chemical shift perturbations. (F) 2D NOESY spectra overlay of unbound Loc1p peptide (red) and dsRNA-bound Loc1p (black). In the free state, the Loc1p peptide is flexible, as only intra-residual and sequential HN–Hα1 NOE cross-peaks are observed. Upon binding to dsRNA, additional NOEs can be observed, especially nonsequential and inter-residual NOEs. The dashed lines indicate strips from the HN region of indicated residues to other protons. The purple circle indicates the strong overlap of positively charged residues (K4, K5, K8, R14), which prevents full assignment of peptide side chains. Secondary chemical shifts (below the spectrum) clearly show that no secondary structure is formed upon RNA binding. (G) TAR annealing assay with radioactively labeled TAR(+) and unlabeled minus strand demonstrates that concatenation of the most N-terminal Loc1p motif is sufficient to mimic the annealing activity of wild-type Loc1p in vitro. Both strands were incubated with increasing concentrations of wild-type Loc1p, Loc1p (1–20)10, Loc1p (1–20)2, Loc1p (1–20) or buffer (negative control: NC) at 37°C. Heat-induced displacement at 70°C and in the absence of Loc1p served as a positive control (PC). Formation of dsTAR was analyzed by 8% native TBE PAGE. Wild-type Loc1p has an annealing activity comparable to an artificial protein bearing 10 Loc1p motifs [Loc1p (1–20)10]. (H) Comparison of annealing activities of sequences bearing 1, 2 and 10 Loc1p motifs. For Loc1p (1–20), annealing activity was not determined because even with 50 μM no saturation was reached. According to the intensities of annealed dsTAR, Loc1p (1–20)2 showed an estimated 50–100-fold higher activity than Loc1p (1–20). Furthermore Loc1p (1–20)10 showed an over 50-fold higher annealing activity than Loc1p (1–20)2. Since wild-type Loc1p showed even better reactivity that exceeded the limits of this assay, no Km could be derived. Shown are quantifications from three independent experiments.