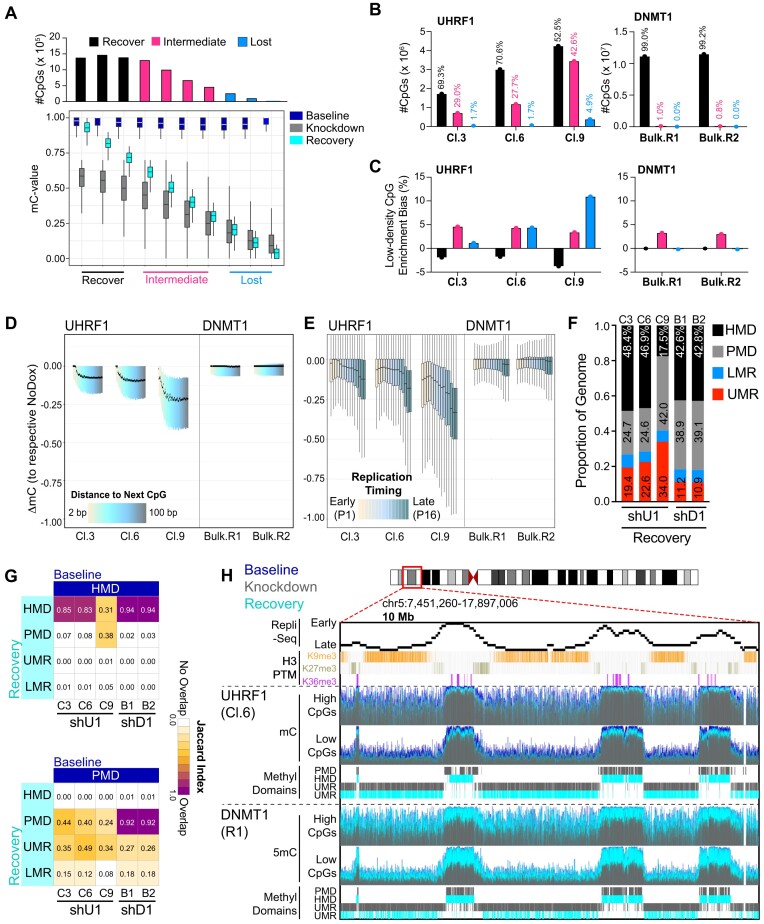
Figure 3.
Recovery of low-density CpG methylation requires UHRF1. (A) Recovery analysis strategy for classifying recovery dynamics of significantly hypomethylated CpGs (from Supplementary Figure S1D) across shUHRF1 and shDNMT1 (WGBS) samples. shUHRF1 Clone 9 is presented to illustrate recovery dynamic binning analysis. DNA methylation recovery is determined by calculating the ΔmC of Day 28 samples (Recovery) to the respective Baseline (NoDox) samples. DNA methylation recovery measurements were then binned into three groups, where CpGs with recovery values (|ΔmCRecovery|*100) < 30% are considered lost, > 70% are considered recovered, and between 30 and 70% are considered intermediate recovered. (B) Distribution of recovery categories among shUHRF1 and shDNMT1 samples (WGBS). Legend from 3A applies. (C) Hypergeometric analysis for enrichment of low-density CpGs across recovery categories for each shUHRF1 and shDNMT1 sample (WGBS) from 3B. (D) Boxplots for change in methylation recovery (ΔmCRecovery) of all highly methylated CpGs (mCBaseline 0.85) from WGBS of each indicated sample across CpG binning by ‘Distance to the Next CpG’ [color bar (2–100 bp)]. Whiskers were removed for figure clarity. (E) Boxplots for change in methylation recovery (ΔmCRecovery) of all highly methylated CpGs (mCBaseline
0.85) from WGBS of each indicated sample across CpG binning by ‘Distance to the Next CpG’ [color bar (2–100 bp)]. Whiskers were removed for figure clarity. (E) Boxplots for change in methylation recovery (ΔmCRecovery) of all highly methylated CpGs (mCBaseline 0.85) from WGBS of each indicated sample across replication timing phases. (F) Proportional coverage of the genome of called Methylation Domains in the Recovery stage for each shUHRF1 and shDNMT1 sample (WGBS). (G) Overlap analysis of called Methylation Domains from Baseline and Recovery methylomes across shUHRF1 and shDNMT1 WGBS samples. The Jaccard Index measures the extent of the overlap with 0 being no overlap and 1 being complete overlap. (H) Browser shot of 10 Mb region on Chr5 demonstrating the observations made from WGBS analysis of the Recovery methylome in shUHRF1 Cl.6 and shDNMT1 replicate 1 samples integrated with Repli-Seq, histone PTMs, and locality of called Methylation Domains. See alsoSupplementary Figure S3.
0.85) from WGBS of each indicated sample across replication timing phases. (F) Proportional coverage of the genome of called Methylation Domains in the Recovery stage for each shUHRF1 and shDNMT1 sample (WGBS). (G) Overlap analysis of called Methylation Domains from Baseline and Recovery methylomes across shUHRF1 and shDNMT1 WGBS samples. The Jaccard Index measures the extent of the overlap with 0 being no overlap and 1 being complete overlap. (H) Browser shot of 10 Mb region on Chr5 demonstrating the observations made from WGBS analysis of the Recovery methylome in shUHRF1 Cl.6 and shDNMT1 replicate 1 samples integrated with Repli-Seq, histone PTMs, and locality of called Methylation Domains. See alsoSupplementary Figure S3.