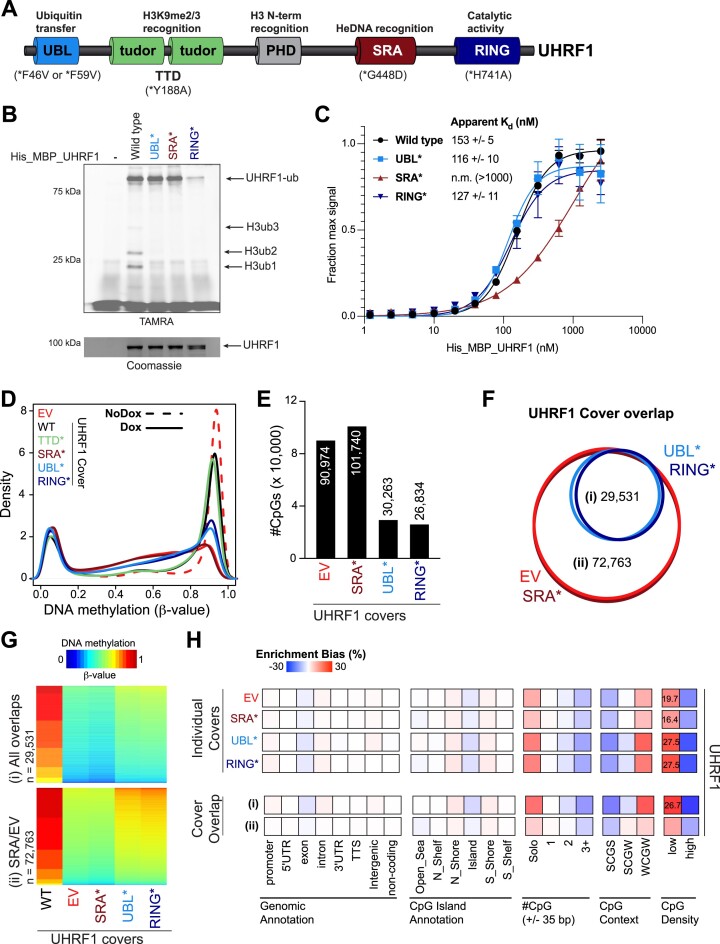
Figure 4.
UHRF1 ubiquitin ligase activity is required for the maintenance of low-density CpG methylation. (A) Schematic of UHRF1 protein domains and mutations made to the UHRF1 transgene covers. (B) In vitro ubiquitin ligase activity assays with recombinant WT UHRF1 or the indicated domain loss-of-function mutants and unmodified nucleosomes with 26 bp linker DNA as substrate. (C) In vitro Alpha-screen proximity assays measuring interactions between unmodified nucleosomes with 26 bp linker DNA and recombinant recombinant WT UHRF1 or the indicated domain loss-of-function mutants. (D) Density plot for CpG probe distribution across DNA methylation levels [β-value: 0 (unmethylated) to 1 (methylated)] for WT and mutant(*) UHRF1 transgene covers. (E) Bar graph of number of hypomethylated CpGs (EPIC array) for each UHRF1 cover (β-valueWT_cover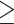 0.85; Δβ-valuemutant_covers
0.85; Δβ-valuemutant_covers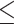 −0.3). (F) Venn diagram demonstrating overlap of hypomethylated CpGs from UHRF1 cover experiments. (G) Heatmap of DNA methylation (β-value) values for overlapping hypomethylated CpGs from the (i) Ubiquitin mutant covers and (ii) EV and SRA mutant covers. (H) Hypergeometric analysis of hypomethylated CpGs from UHRF1 cover experiments. Positive values indicate significant overrepresentation for hypomethylation of the feature (genomic annotation, CpG island annotation, etc.) and negative values indicate significant underrepresentation. Enrichment bias values are provided for the most significant positive enrichments. See alsoSupplementary Figure S4.
−0.3). (F) Venn diagram demonstrating overlap of hypomethylated CpGs from UHRF1 cover experiments. (G) Heatmap of DNA methylation (β-value) values for overlapping hypomethylated CpGs from the (i) Ubiquitin mutant covers and (ii) EV and SRA mutant covers. (H) Hypergeometric analysis of hypomethylated CpGs from UHRF1 cover experiments. Positive values indicate significant overrepresentation for hypomethylation of the feature (genomic annotation, CpG island annotation, etc.) and negative values indicate significant underrepresentation. Enrichment bias values are provided for the most significant positive enrichments. See alsoSupplementary Figure S4.