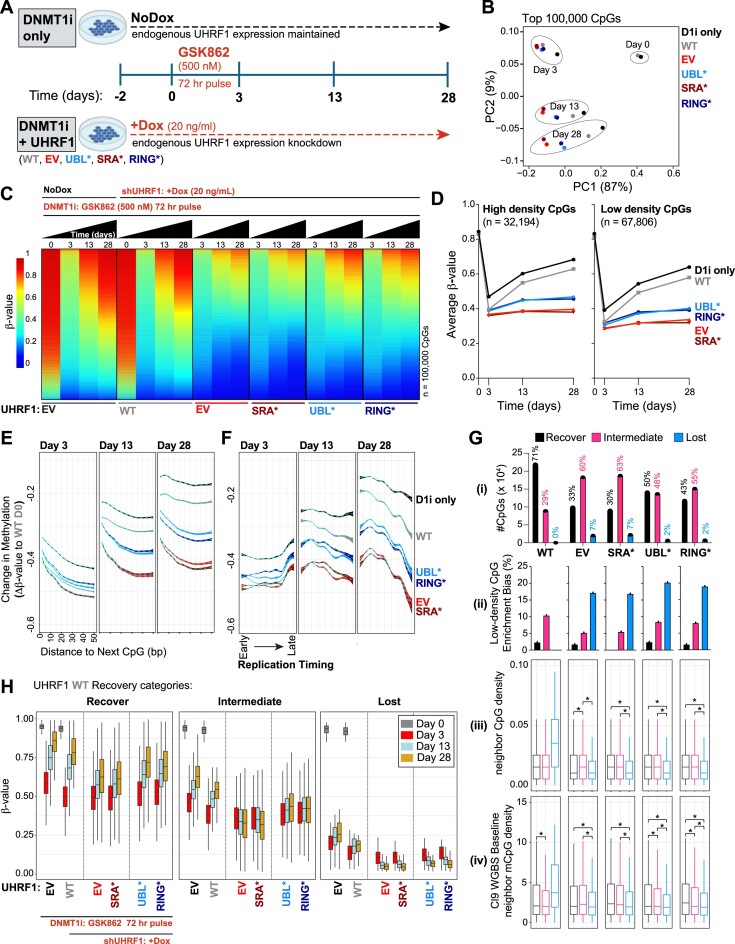
Figure 5.
UHRF1 ubiquitin ligase activity is required for recovery of low-density CpG methylation. (A) Schematic of treatment paradigm in HCT116 dox-inducible shUHRF1 Clone 9 cells with UHRF1 transgene covers (used in experiments presented in Figure 4). To inhibit DNMT1 activity, all cell lines were treated with a single-dose of GSK-3484862 (GSK862) (500 nM) for 72 h. For the DNMT1i only control (D1i only), cells were not treated with dox in order to maintain expression of endogenous UHRF1. DNMT1i + UHRF1 cover lines were pretreated with dox (20 ng/ml) for 48 h prior to GSK862 to remove expression of endogenous UHRF1 and measure depletion and recovery of DNA methylation in the presence of the UHRF1 transgene cover. Dox treatment (20 ng/ml) was maintained throughout the duration of the experiment (28 days) to maintain knockdown of endogenous UHRF1. Created with BioRender.com (B) Principal components analysis of the top 100 000 most variably methylated CpGs (EPIC array) across all samples and time-points. (C) Heatmaps of the top 100 000 most differentially methylated CpGs (comparison: Day 28 UBL*/RING* versus Day 28 D1i only/D1i + WT cover). CpGs are ranked from highest to lowest DNA methylation for the Day 28 WT cover cells. (D) Average DNA methylation of top 100 000 most differentially methylated high-density CpGs (left) and low-density CpGs (right) from C across all time-points. (E) Average loss of DNA methylation for all highly methylated CpGs (β-value(WTcover_Day0)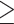 0.85) across bins of decreasing CpG density. Dotted line indicates the average Δβ-value as a function of distance. Boundaries indicate 95% confidence intervals. (F) Average loss of DNA methylation for all highly methylated CpGs (β-value(WTcover_Day0)
0.85) across bins of decreasing CpG density. Dotted line indicates the average Δβ-value as a function of distance. Boundaries indicate 95% confidence intervals. (F) Average loss of DNA methylation for all highly methylated CpGs (β-value(WTcover_Day0)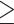 0.85) across replication timing phases. Dotted line indicates the average Δβ-value as a function of replication timing. Boundaries indicate 95% confidence intervals. (G) Characterization of recovery dynamic categories across UHRF1 transgene cover experiments: (i) Number of significantly hypomethylated CpGs (β-value(WTcover_Day0)
0.85) across replication timing phases. Dotted line indicates the average Δβ-value as a function of replication timing. Boundaries indicate 95% confidence intervals. (G) Characterization of recovery dynamic categories across UHRF1 transgene cover experiments: (i) Number of significantly hypomethylated CpGs (β-value(WTcover_Day0)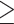 0.85; Δβ-value(Day3-WTcover_Day0)
0.85; Δβ-value(Day3-WTcover_Day0)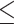 −0.3) that demonstrate indicated recovery dynamics among the UHRF1 cover transgene experiments. (ii) Hypergeometric analysis for enrichment of low-density CpGs across recovery dynamic categories for each UHRF1 transgene cover experiment. (iii) Boxplots of neighboring CpG density (−/+ 100 bp flanking CpG of interest). *pval < 2.2e-16 by one-sided Mann–Whitney U-test. (iv) Boxplots of neighboring methylated CpG density (values derived from shUHRF1 Cl.9 baseline WGBS; −/+ 100 bp flanking CpG of interest). *P-value < 2.2e-16 by one-sided Mann–Whitney U-test. (H) DNA methylation distributions across the time-course for CpGs that recover with UHRF1 WT (left), intermediately recover with UHRF1 WT (middle), or are lost (right). DNA methylation values among the EV and UHRF1 mutant covers for CpGs in the designated UHRF1 WT recovery bins demonstrate which CpGs require UHRF1 for DNA methylation recovery. See also Supplementary Figure S5.
−0.3) that demonstrate indicated recovery dynamics among the UHRF1 cover transgene experiments. (ii) Hypergeometric analysis for enrichment of low-density CpGs across recovery dynamic categories for each UHRF1 transgene cover experiment. (iii) Boxplots of neighboring CpG density (−/+ 100 bp flanking CpG of interest). *pval < 2.2e-16 by one-sided Mann–Whitney U-test. (iv) Boxplots of neighboring methylated CpG density (values derived from shUHRF1 Cl.9 baseline WGBS; −/+ 100 bp flanking CpG of interest). *P-value < 2.2e-16 by one-sided Mann–Whitney U-test. (H) DNA methylation distributions across the time-course for CpGs that recover with UHRF1 WT (left), intermediately recover with UHRF1 WT (middle), or are lost (right). DNA methylation values among the EV and UHRF1 mutant covers for CpGs in the designated UHRF1 WT recovery bins demonstrate which CpGs require UHRF1 for DNA methylation recovery. See also Supplementary Figure S5.