Abstract
Background
Due to limited treatment options, cutaneous warts caused by human papillomavirus (HPV) remain a significant clinical challenge. Furthermore, the genetic susceptibility and molecular basis of viral warts are not yet fully understood.
Methods
We utilized a multi-omics integration approach, encompassing genome-wide association study (GWAS) meta-analysis, summary data-based Mendelian randomization (SMR) analysis, and transcriptomic validation using the GSE136347 dataset. Differential gene expression (DEG) analysis was conducted to identify significant changes in gene expression between wart tissues and healthy skin.
Results
Our analyses revealed five genetic susceptibility genes associated with cutaneous warts, with RARA showing significant differential expression in wart tissues. Co-expression analysis indicated that RARA may regulate apoptosis through interactions with BAX, a pro-apoptotic gene. Additionally, functional annotation via Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses highlighted key biological processes and pathways involved in wart pathogenesis.
Conclusion
This study identifies RARA as a pivotal regulator in the molecular pathology of cutaneous warts and a promising therapeutic target. RA-based therapies could offer effective and less invasive alternatives for wart treatment. Future investigations should refine the molecular role of RARA to optimize clinical interventions.
Keywords: cutaneous warts, GWAS-meta, multi-omics, RARA, retinoic acid
Introduction
Cutaneous warts are caused by infection with human papillomavirus (HPV) and can be classified into several types based on the site of infection, including common warts, flat warts, plantar warts, and genital warts.1 As a prevalent clinical condition, studies indicate that 5–30% of children and adults are affected by warts.2,3 Certain types, such as plantar warts, are often neglected during early stages, with patients seeking medical attention only after the lesions increase in number or spread.4 While some warts may resolve spontaneously within months or years, cases persisting for 5 to 10 years are not uncommon. Furthermore, warts carry the risk of direct or indirect transmission to others, highlighting the importance of early and effective treatment.
Despite a range of treatment options, managing warts remains challenging due to the absence of HPV-specific antiviral drugs. Current therapies fall into six main categories: destructive treatments, virucidal agents, antiproliferative drugs, immunotherapies, complementary and alternative therapies, and other approaches. Each comes with limitations. For instance, destructive treatments like salicylic acid or cryotherapy often cause pain, scarring, and pigmentation changes. Similarly, virucidal agents and antiproliferative drugs, such as bleomycin and 5-fluorouracil, are associated with adverse effects like local tissue necrosis and systemic toxicity. Immunotherapies, including imiquimod, can lead to erythema or depigmentation, while complementary therapies like herbal treatments often have unproven efficacy. These limitations highlight the urgent need for safer and more effective treatment strategies.
Recent advances in genomics and bioinformatics have provided unprecedented opportunities to uncover the genetic predispositions underlying various diseases.5,6 Genome-wide association studies (GWAS) play a crucial role in identifying genetic susceptibility loci by analyzing millions of genetic variants across large populations to detect statistically significant associations with specific traits or diseases.7 This approach has provided insights into conditions ranging from cancer to autoimmune disorders, paving the way for personalized medicine. For example, genetic findings have guided the development of targeted therapies, such as using specific inhibitors for genetically driven cancers.8 In 2021, two-thirds of FDA-approved drugs had genetic evidence supporting their efficacy,9 underscoring the increased likelihood of success for genetically supported drug targets in clinical trials.10 These studies offer valuable insights into the genetic architecture of complex diseases, by highlighting previously unknown molecular players involved in pathogenesis. However, individual GWAS can sometimes suffer from insufficient power due to limited sample sizes, resulting in false negatives or underrepresentation of weaker genetic signals.11
To address these limitations, GWAS-meta analysis integrates data from multiple studies, enhancing statistical power and the ability to detect smaller genetic effects. By identifying robust susceptibility loci, this approach offers the potential to uncover novel molecular pathways involved in disease pathogenesis.12 Integrating GWAS findings with transcriptomic data further validates the biological relevance of genetic variants by examining their functional expression in specific tissues. Such multi-omics approaches have proven valuable for bridging the gap between genetic discoveries and clinical applications, ultimately aiding in the identification of therapeutic targets.13
In this study, our goal is to elucidate the genetic susceptibility loci and molecular mechanisms of cutaneous warts through multi-omics integration and to explore potential therapeutic targets. We performed a GWAS-meta analysis using wart-related genome-wide data from FinnGen, GWAS Catalog, and UK Biobank, followed by the identification of candidate susceptibility genes through FUMA and MAGMA tools. To further explore causal relationships, we employed summary-based Mendelian randomization (SMR), utilizing eQTLgen as the discovery dataset and GTEx as the validation dataset. To validate the expression of these candidate genes in skin tissue, we conducted transcriptomic analyses of wart samples from the GEO database and explored the molecular pathways involved. This integrated approach offers new insights into the mechanisms of wart development and helps identify potential therapeutic targets.
Materials and Methods
Study Design
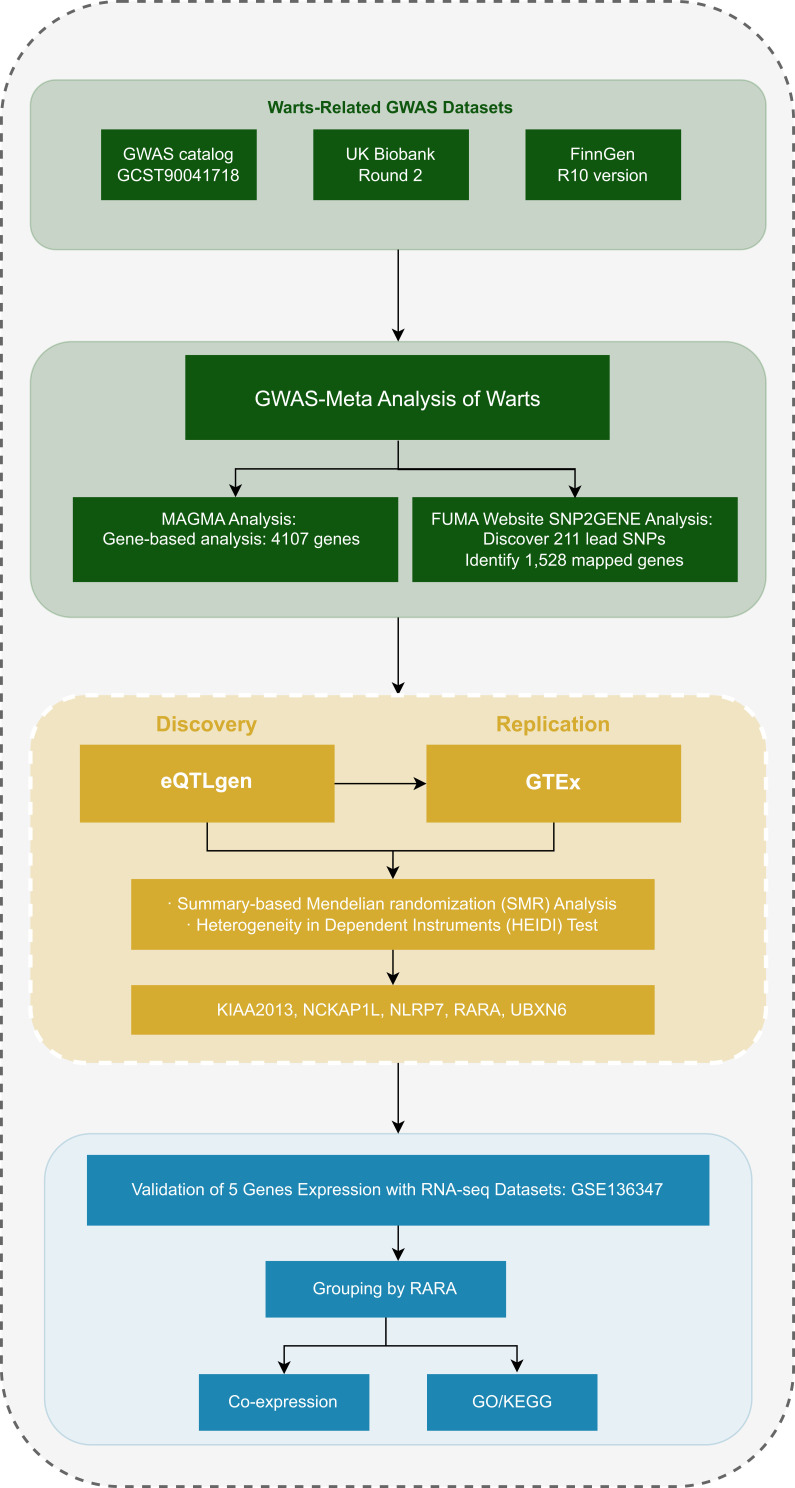
This study primarily includes GWAS-meta analysis, SMR analysis, and RNA-seq analysis. The research workflow is presented in Figure 1. Refer to Supplementary Table S1 for detailed information regarding the data.
Figure 1.
Study design overview.
GWAS-Meta Analysis
We integrated multiple warts GWAS datasets, including those from the GWAS Catalog, UK Biobank, and FinnGen, comprising a total of 1,217,097 individuals and 22,447,090 SNPs. A GWAS-meta analysis was conducted to identify genetic loci associated with warts. METAL soft14 (https://genome.sph.umich.edu/wiki/METAL_Documentation) was used for performing GWAS meta-analysis. A sample size weighted meta-analysis of Z scores (Stouffer’s method) was employed. This method converts the observed effect direction and P-value in each study into a signed Z-score: A very negative Z-score indicates a small P-value and that the allele is associated with a lower disease risk or quantitative trait level. A large Z-score indicates a small P-value and that the allele is associated with a higher disease risk or quantitative trait level. The Z-scores for each allele were combined across samples using a weighted sum, where the weights are proportional to the square root of the sample size in each study.15
FUMA Analysis
We utilized the Functional Mapping and Annotation of Genome-Wide Association Studies (FUMA) platform (https://fuma.ctglab.nl/snp2gene) to annotate and map single nucleotide polymorphisms (SNPs) to genes. FUMA is an integrative web-based tool that links GWAS results to biological functions by leveraging multiple data repositories, including eQTLs, chromatin interactions, and tissue-specific gene expression profiles. This approach allows for the prioritization of candidate genes and identification of relevant pathways associated with specific phenotypes. In our analysis, all parameters were set to the default thresholds provided by the FUMA website to ensure standardized and reproducible results. This tool facilitated the identification of lead SNPs, gene-level associations, and potential regulatory elements linked to the genetic architecture of warts.
MAGMA Analysis
We performed Multi-marker Analysis of Genomic Annotation (MAGMA) analysis using MAGMA v1.10 to identify genes genetically associated with warts. MAGMA is a widely used tool designed for gene-based analysis, which aggregates the effects of multiple SNPs across the genome to test for their collective association with a given trait.16,17 This approach is particularly useful for uncovering functional genes or biological modules, such as regulatory pathways, that may not be detected through individual SNP-based methods alone. It allows for the identification of genes influenced by multiple small-effect SNPs,18 thus capturing more subtle, polygenic contributions to complex traits like warts. Furthermore, MAGMA integrates functional annotations, such as expression profiles and chromatin states, enabling pathway level enrichment analysis to identify key biological processes involved in disease pathogenesis. This analysis provides insights into the genetic architecture and biological underpinnings of warts, supporting the prioritization of candidate genes for further investigation.
SMR Analysis
To explore the genetic susceptibility underlying warts, we conducted Summary-data-based Mendelian Randomization (SMR) analysis using eQTL data from the eQTLGen19 consortium and GTEx V8.20 SMR analysis was performed with SMR 1.3.1 software, which integrates GWAS and eQTL data to investigate whether the association between SNPs and a trait is mediated through gene expression. Specifically, we utilized SMR-format files to evaluate the overlap between expression quantitative trait loci (eQTLs) and warts-related SNPs, aiming to identify genetic susceptibility genes whose expression may contribute to wart development.
We established discovery and validation datasets, with eQTLGen serving as the discovery set and GTEx as the validation set, ensuring the robustness and reproducibility of our findings across independent datasets. The SMR framework provides a powerful tool for integrating genetic association data with transcriptomic information, helping to reveal key molecular mechanisms involved in disease onset and progression. A crucial element of the SMR approach is the HEIDI (Heterogeneity in Dependent Instruments) test, which assesses whether the association between SNPs and gene expression is due to pleiotropy or linkage disequilibrium (LD). If the HEIDI test confirms that the observed association is not driven by LD,21,22 it strengthens the evidence for the gene’s role as a genetic susceptibility factor for the disease. Together, the SMR analysis and HEIDI test offer valuable insights into the molecular pathways underlying warts, aiding in the identification of potential therapeutic targets for future research.
Validation Using RNA-Seq Data
To validate the expression of genetic susceptibility genes in wart tissue, we utilized the GSE136347 dataset from the GEO database, containing transcriptomic data from human HPV-induced warts and healthy skin. Differential gene expression (DEG) analysis was performed using the Limma package (version 3.58.1) in R, employing a threshold of |LogFC| > 1 and an adjusted p-value < 0.05. This facilitated precise identification of genes that are significantly differentially expressed between wart tissues and healthy skin, ensuring statistical robustness. This DEG analysis was critical for confirming whether the previously identified genetic susceptibility genes exhibit significant differential expression in HPV-induced warts.
Subsequently, we stratified the wart samples based on the median expression levels of differentially expressed genes. We then conducted Gene Ontology (GO) analysis to annotate these genes in terms of their biological processes, cellular components, and molecular functions, thereby illuminating the roles these genes may play in cellular dynamics relevant to wart pathogenesis. Additionally, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was employed to identify enriched pathways, which provided insight into relevant biological and signaling pathways associated with HPV-induced warts.
To further examine the molecular interactions involving the identified susceptibility genes, co-expression analysis was conducted. This analysis allowed us to explore potential regulatory relationships, revealing key genes that may be co-regulated or functionally linked with the identified genetic susceptibility genes in the context of HPV-induced warts. This integrative approach highlights the biological significance of these genes in wart development and their potential as therapeutic targets.
Results
Results of FUMA, MAGMA and SMR Analysis
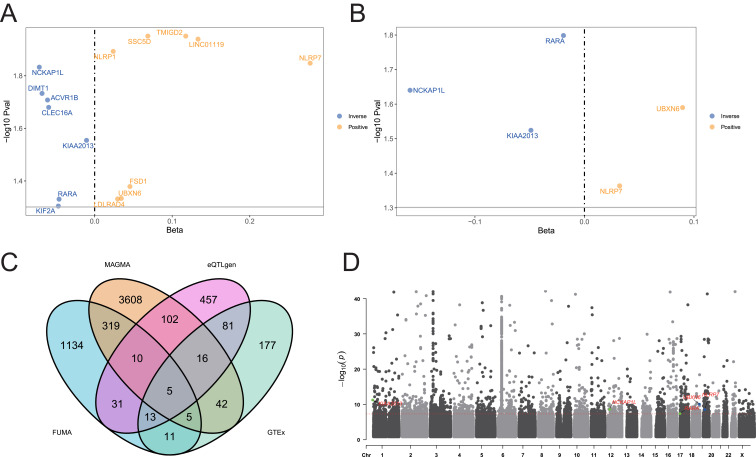
The GWAS-meta analysis generated novel GWAS data (Supplementary Figure 1A). Using the FUMA platform, we annotated SNP loci and identified a total of 211 SNPs (Supplementary Table S2), mapped to 1528 genes (Supplementary Table S3). The results of the MAGMA analysis are provided in Supplementary Table S4. By intersecting the findings from FUMA and MAGMA, we identified 339 overlapping genes (Supplementary Figure 1B). Of these, 15 genes were found to be closely associated with warts through SMR analysis based on the eQTLGen dataset. Further validation using SMR analysis (Figure 2A) with the GTEx (Figure 2B) dataset confirmed the association of five of these genes with warts. Thus, through systematic filtering, we identified five genetic susceptibility genes (RARA, UBXN6, KIAA2013, NLRP7, NCKAP1L) associated with warts at the genome-wide level (Figure 2C). The genomic distribution of these five genes is shown in Figure 2D. Detailed results of the SMR analysis are provided in Supplementary Table S5-S6.
Figure 2.
(A) Results of SMR analysis based on the eQTLgen database, identifying genes with a significant association with warts. Yellow bars indicate genes with a positive beta value (Beta > 0), suggesting a potential positive association with disease progression, while blue bars represent genes with a negative beta value (Beta < 0), indicating a potential inhibitory role in disease progression. (B) Results of SMR analysis based on the GTEx database, further validating five genes with significant associations. Similarly, yellow bars denote genes with positive beta values, implying potential promotive effects on disease, while blue bars represent genes with negative beta values, indicating potential suppressive effects. (C) Final selection of 5 genetic susceptibility genes for warts after multi-step filtering through integrated analysis of eQTLgen and GTEx databases, highlighting the robustness of these candidates. (D) Genomic distribution of the 5 identified genetic susceptibility genes, visualized across the genome to illustrate their locations on different chromosomes, providing a comprehensive view of their genetic context.
Results of RNA-Seq Analysis
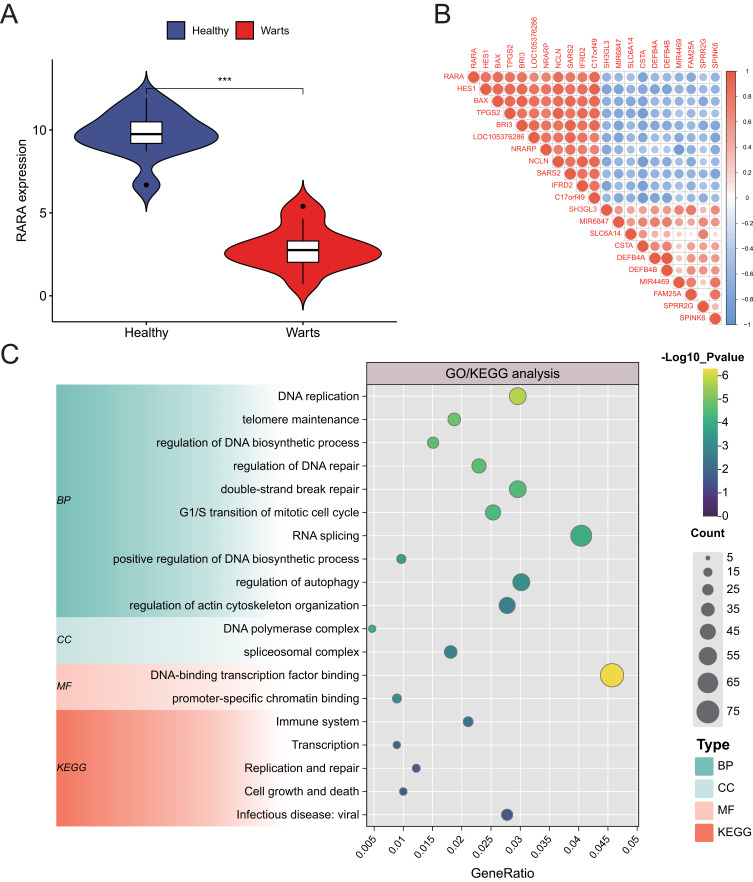
Principal Component Analysis (PCA) clustering was initially performed on the RNA-seq data from the GSE136347 dataset. The results demonstrated high data quality, with clear separation between healthy skin and wart samples (Supplementary Figure 2A). Differential expression analysis of the five genetic susceptibility genes identified in Results of FUMA, MAGMA and SMR Analysis revealed that RARA exhibited significant differential expression between the two groups (Supplementary Figure 2B and Figure 3A).Further analysis grouped the wart tissue samples into high- and low-expression groups based on the median expression level of RARA. Co-expression analysis indicated that in the RARA high-expression group, the apoptosis-related protein BAX was also highly expressed (Figure 3B), suggesting a potential association between RARA and BAX. Gene Ontology (GO) and KEGG pathway enrichment analyses highlighted that differentially expressed genes between the RARA high- and low-expression groups play crucial roles in DNA metabolism, cell cycle regulation, and transcriptional control. These genes may also be associated with viral infection and immune responses (Figure 3C). The detailed results of the RNA-seq analysis can be found in Supplementary Table S7.
Figure 3.
(A) Violin plot illustrating RARA expression levels in wart tissue compared to control samples, further confirming its differential expression and potential involvement in wart pathogenesis. The widened areas of the plot indicate higher sample density, providing a clear view of expression variability. *** indicates P < 0.001, representing results with extremely significant statistical difference. (B) Co-expression analysis stratified by median RARA expression, showing increased BAX expression in the high-RARA group. This suggests a potential mechanistic link between RARA and apoptosis-related genes, which may contribute to the apoptotic clearance of HPV-infected cells. The stratification allows for a focused comparison of gene interactions within biologically relevant expression thresholds. (C) Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of differentially expressed genes (DEGs) between high- and low-RARA expression groups. The analysis highlights enriched biological functions and pathways, including DNA metabolism, cell cycle regulation, transcriptional control, immune response, and viral infection pathways. These findings provide insights into the broader molecular context in which RARA functions, suggesting its potential roles in both host defense mechanisms and HPV-associated processes.
Discussion
This study provides a comprehensive exploration of the genetic architecture and potential molecular mechanisms underlying cutaneous warts by integrating GWAS-meta analysis, SMR analysis, and transcriptomic validation. Leveraging data from large-scale genomic databases and transcriptomic profiling, we identified five genetic susceptibility genes (RARA, UBXN6, KIAA2013, NLRP7, NCKAP1L) linked to warts, with RARA being particularly noteworthy due to its differential expression in HPV-induced wart tissues.
The Role of Retinoic Acid in Regulating RARA Expression
Retinoic acid (RA) is a critical regulator of gene expression, playing essential roles in cellular differentiation, proliferation, and apoptosis, particularly in epithelial tissues. RA exerts its effects primarily through binding to nuclear retinoic acid receptors (RARs), which belong to the nuclear receptor superfamily and include three subtypes: RARα, RARβ, and RARγ. These receptors form heterodimers with retinoid X receptors (RXRs) and act as ligand-dependent transcription factors. Upon activation, these heterodimers bind to specific retinoic acid response elements (RAREs) in the promoter regions of target genes, modulating their expression.23,24 Through platforms such as the Open Targets Platform, RA has been identified as a potent retinoic acid receptor agonist targeting RARα, RARβ, and RARγ, with its mechanism of action directly linked to RAR-mediated transcriptional regulation.
The therapeutic relevance of RA-RARA signaling is well-documented, spanning applications in both cancer25 and epithelial dysfunction-related diseases. For example, RA significantly improved nephrotoxic serum-induced glomerulonephritis in wild-type mice by activating RARα, as demonstrated by its inhibition of podocyte proliferation and induction of differentiation.26 These effects were notably diminished in podocyte-specific Rara knockout mice, highlighting the specificity of RA’s action via RARα (Retinoic acid improves nephrotoxic serum-induced glomerulonephritis through activation of podocyte retinoic acid receptor α). This pathway underscores the potential of RA-RARA interaction in modulating epithelial cell behavior.
In the context of cutaneous warts, RA-RARA signaling may play a similar role by regulating keratinocyte proliferation and differentiation, processes central to HPV-induced lesion development. Targeting this pathway could represent a promising therapeutic strategy, offering insights into new approaches for effective wart treatment.
Significance of RARA in Wart Pathogenesis
Among the five genes identified, RARA (retinoic acid receptor alpha) stood out due to its significant differential expression in wart tissues compared to healthy skin. Retinoic acid, the ligand for RARA, modulates crucial cellular processes such as proliferation, differentiation, and apoptosis. The observed upregulation of RARA in wart samples, along with its co-expression with BAX, a pro-apoptotic protein, suggests a potential role for RARA in regulating cell death pathways involved in wart pathogenesis. This aligns with prior research emphasizing retinoic acid’s capacity to modulate immune responses and promote apoptosis in HPV-infected cells. The high expression of BAX in the RARA high-expression group supports the hypothesis that RARA might enhance the apoptotic response, potentially limiting the spread of HPV-infected cells.
Retinoic acid has a well-established role in dermatological treatments, marking the third therapeutic revolution in dermatology due to its nuclear receptor-mediated biological activities.27 These activities include anti-proliferative effects, differentiation regulation, immune modulation, sebum secretion suppression, and anti-aging effects. Its use in conditions such as psoriasis, acne, ichthyosis, and mycosis fungoides demonstrates its broad applicability.28 Prior studies have shown that retinoic acid is effective in treating skin warts with good safety and less pain compared to physical treatments.29 However, the specific mechanisms through which retinoic acid exerts its therapeutic effects on warts remain unclear, making this study’s focus on RARA particularly significant.
Role of Other Identified Genes
The other identified genes (UBXN6, KIAA2013, NLRP7, and NCKAP1L) also provide valuable insights into the molecular pathways involved in wart development. Although they did not exhibit significant differential expression in RNA-seq analysis, their association with warts in GWAS-meta and SMR analyses underscores their potential involvement in disease pathogenesis. For instance, UBXN6’s role in protein degradation pathways could influence viral protein turnover and immune evasion.30 NLRP7, known for its role in inflammasome formation, hints at a possible link between innate immunity and wart susceptibility.31
Integration of Multi-Omics Approaches
The integration of GWAS-meta, SMR, and RNA-seq analyses strengthens the robustness of our findings. The GWAS-meta analysis enhanced statistical power and reliability, revealing novel SNP loci associated with warts. Candidate genes were identified and prioritized through functional annotation using tools such as FUMA and MAGMA. The SMR analysis further validated these findings by linking genetic loci to gene expression, with HEIDI testing to rule out linkage disequilibrium as a confounding factor. The RNA-seq validation, utilizing data from the GSE136347 dataset, provided essential biological context, affirming RARA’s significant role in HPV-induced warts.
Implications for Treatment and Future Research
The identification of RARA as a key susceptibility gene has therapeutic implications. Retinoic acid and its derivatives are widely used in dermatology, and our findings suggest that targeting the RARA pathway could be an effective strategy for wart treatment. The association between RARA and apoptosis-related genes, such as BAX, indicates that retinoic acid treatment may promote the clearance of HPV-infected cells through apoptotic mechanisms.
Future studies should explore the therapeutic potential of modulating RARA and related pathways in wart treatment. In vitro and in vivo studies focusing on the mechanistic role of RARA in HPV-infected keratinocytes could elucidate how it influences viral replication and host immune responses. Additionally, further research is needed to investigate the roles of UBXN6, KIAA2013, NLRP7, and NCKAP1L in wart pathogenesis to understand their contributions to disease susceptibility and progression.
Limitations
This study has some limitations. While the multi-omics approach provided robust candidate gene identification, functional validation was restricted to existing RNA-seq datasets. Direct experimental studies involving the manipulation of these genes in HPV models are essential to confirm their roles in wart pathogenesis and should be pursued in future research. Moreover, while the GWAS-meta analysis included data from diverse populations, potential population-specific genetic factors may not have been fully captured. In addition, the genomics content presented in this study may be challenging for clinicians due to its complexity. To mitigate this, we have included more detailed explanations of the bioinformatics terms (eg, FUMA, MAGMA, SMR) in Supplementary Table S8 to support understanding, particularly for dermatologists.
Conclusion
This study underscores the pivotal role of retinoic acid (RA) and its receptor RARA in the pathogenesis and treatment of cutaneous warts. Through multi-omics integration, including GWAS-meta analysis, SMR analysis, and transcriptomic validation, we identified RARA as a key susceptibility gene, highlighting its potential as a therapeutic target. The co-expression of RARA with apoptosis-related genes such as BAX suggests that RA may promote the apoptotic clearance of HPV-infected cells, consistent with its known dermatological activities. Our findings support RA as a safer and less painful alternative to conventional treatments for warts. Based on these results, we recommend further prospective clinical trials to assess RA’s therapeutic effects on persistent warts. These trials could compare cryotherapy, RA treatment, and a combination of both, with primary endpoints focusing on the reduction rate of plantar warts at 8 weeks. Secondary endpoints would include the time to complete wart resolution, adverse event incidence, and recurrence rates at 4, 8, and 12 weeks post-treatment. Additionally, baseline and post-treatment levels of RARA and BAX expression could be measured to explore the association between RARA and apoptosis in wart tissues, further elucidating RA’s therapeutic mechanism. In summary, this research provides a foundation for advancing RA-based treatments, with the potential to improve clinical outcomes and reduce recurrence in cutaneous wart management.
Ethics Statement
This study is exempt from ethical review as per Article 32 of the Measures for Ethical Review of Life Science and Medical Research Involving Human Beings (National Science and Technology Ethics Committee, China). The exemption is based on the use of non-harmful, non-sensitive data from open, legal databases.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Mlynarczyk-Bonikowska B, Rudnicka L. HPV infections—classification, pathogenesis, and potential new therapies. Int J Mol Sci. 2024;25(14):7616. doi: 10.3390/ijms25147616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Haalen FM, Bruggink SC, Gussekloo J, Assendelft WJJ, A. H EJ. Warts in primary schoolchildren: prevalence and relation with environmental factors. Br J Dermatol. 2009;161(1):148–152. doi: 10.1111/j.1365-2133.2009.09160.x [DOI] [PubMed] [Google Scholar]
- 3.Kyriakis K, Pagana G, Michailides C, Emmanuelides S, Palamaras I, Terzoudi S. Lifetime prevalence fluctuations of common and plane viral warts. J Eur Acad Dermatol Venereol JEADV. 2007;21(2):260–262. doi: 10.1111/j.1468-3083.2006.01833.x [DOI] [PubMed] [Google Scholar]
- 4.Huang K, Li M, Xiao Y, et al. The application of medical scale in the treatment of plantar warts: analysis and prospect. J Dermatol Treat. 2022;33(2):637–642. doi: 10.1080/09546634.2020.1781757 [DOI] [PubMed] [Google Scholar]
- 5.Sommariva E, Bellin M, Di Resta C. Advance in genomics of rare genetic diseases. Biomolecules. 2023;13(10):1441. doi: 10.3390/biom13101441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abu-Elmagd M, Assidi M, Alrefaei AF, Rebai A. Editorial: advances in genomic and genetic tools, and their applications for understanding embryonic development and human diseases. Front Cell Dev Biol. 2022;10:1016400. doi: 10.3389/fcell.2022.1016400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao X, Wang X, Zhang S, Sha Q. Gene-based association tests using GWAS summary statistics and incorporating eQTL. Sci Rep. 2022;12(1):3553. doi: 10.1038/s41598-022-07465-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaccaro K, Allen J, Whitfield TW, et al. Targeted therapies prime oncogene-driven lung cancers for macrophage-mediated destruction. J Clin Invest. 2024;134(9):e169315. doi: 10.1172/JCI169315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ochoa D, Karim M, Ghoussaini M, Hulcoop DG, McDonagh EM, Dunham I. Human genetics evidence supports two-thirds of the 2021 FDA-approved drugs. Nat Rev Drug Discov. 2022;21(8). doi: 10.1038/d41573-022-00120-3 [DOI] [PubMed] [Google Scholar]
- 10.Trajanoska K, Bhérer C, Taliun D, Zhou S, Richards JB, Mooser V. From target discovery to clinical drug development with human genetics. Nature. 2023;620(7975):737–745. doi: 10.1038/s41586-023-06388-8 [DOI] [PubMed] [Google Scholar]
- 11.Jeng XJ, Hu Y, Sun Q, Li Y. Weak signal inclusion under dependence and applications in genome-wide association study. Ann Appl Stat. 2024;18(1). doi: 10.1214/23-AOAS1815 [DOI] [Google Scholar]
- 12.Defo J, Awany D, Ramesar R. From SNP to pathway-based GWAS meta-analysis: do current meta-analysis approaches resolve power and replication in genetic association studies? Brief Bioinform. 2023;24(1):bbac600. doi: 10.1093/bib/bbac600 [DOI] [PubMed] [Google Scholar]
- 13.Liu X, Peng T, Xu M, et al. Spatial multi-omics: deciphering technological landscape of integration of multi-omics and its applications. J Hematol Oncol. 2024;17(1):72. doi: 10.1186/s13045-024-01596-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinforma Oxf Engl. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim HK, Gonçalves VF, Husain MI, et al. Cross-disorder GWAS meta-analysis of endocannabinoid DNA variations in major depressive disorder, bipolar disorder, attention deficit hyperactivity disorder, autism spectrum disorder, and schizophrenia. Psychiatry Res. 2023;330:115563. doi: 10.1016/j.psychres.2023.115563 [DOI] [PubMed] [Google Scholar]
- 16.de Leeuw CA, Neale BM, Heskes T, Posthuma D. The statistical properties of gene-set analysis. Nat Rev Genet. 2016;17(6):353–364. doi: 10.1038/nrg.2016.29 [DOI] [PubMed] [Google Scholar]
- 17.de Leeuw CA, Stringer S, Dekkers IA, Heskes T, Posthuma D. Conditional and interaction gene-set analysis reveals novel functional pathways for blood pressure. Nat Commun. 2018;9(1):3768. doi: 10.1038/s41467-018-06022-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11(4):e1004219. doi: 10.1371/journal.pcbi.1004219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Võsa U, Claringbould A, Westra HJ, et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat Genet. 2021;53(9):1300–1310. doi: 10.1038/s41588-021-00913-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Consortium G, Anand S, Ardlie KG. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369(6509):1318–1330. doi: 10.1126/science.aaz1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Y, Zeng J, Zhang F, et al. Integrative analysis of omics summary data reveals putative mechanisms underlying complex traits. Nat Commun. 2018;9(1):918. doi: 10.1038/s41467-018-03371-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Z, Zhang F, Hu H, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48(5):481–487. doi: 10.1038/ng.3538 [DOI] [PubMed] [Google Scholar]
- 23.Srinivas H, Xia D, Moore NL, et al. Akt phosphorylates and suppresses the transactivation of retinoic acid receptor alpha. Biochem J. 2006;395(3):653–662. doi: 10.1042/BJ20051794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sato Y, Ramalanjaona N, Huet T, et al. The “phantom effect” of the rexinoid LG100754: structural and functional insights. PLoS One. 2010;5(11):e15119. doi: 10.1371/journal.pone.0015119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Masi A, Leboffe L, De Marinis E, et al. Retinoic acid receptors: from molecular mechanisms to cancer therapy. Mol Aspects Med. 2015;41:1–115. doi: 10.1016/j.mam.2014.12.003 [DOI] [PubMed] [Google Scholar]
- 26.Dai Y, Chen A, Liu R, et al. Retinoic acid improves nephrotoxic serum–induced glomerulonephritis through activation of podocyte retinoic acid receptor α. Kidney Int. 2017;92(6):1444–1457. doi: 10.1016/j.kint.2017.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Almeida NR, Conda-Sheridan M. A review of the molecular design and biological activities of RXR agonists. Med Res Rev. 2019;39(4):1372–1397. doi: 10.1002/med.21578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szymański Ł, Skopek R, Palusińska M, et al. Retinoic acid and its derivatives in skin. Cells. 2020;9(12):2660. doi: 10.3390/cells9122660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang S, Xu X, Liu Y, et al. Successful clearance of extensive/recalcitrant cutaneous warts by Acitretin monotherapy: a case series study. Dermatol Ther. 2020;33(3):e13390. doi: 10.1111/dth.13390 [DOI] [PubMed] [Google Scholar]
- 30.Harrison AG, Ketkar H, Wang P. UBX domain protein 6 positively regulates JAK-STAT1/2 signaling. J Immunol. 2022;208(1_Supplement):52.11. doi: 10.4049/jimmunol.208.Supp.52.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jha S, Ting JPY. Inflammasome-associated nucleotide-binding domain, leucine-rich repeat proteins and inflammatory diseases. J Immunol. 2009;183(12):7623–7629. doi: 10.4049/jimmunol.0902425 [DOI] [PMC free article] [PubMed] [Google Scholar]