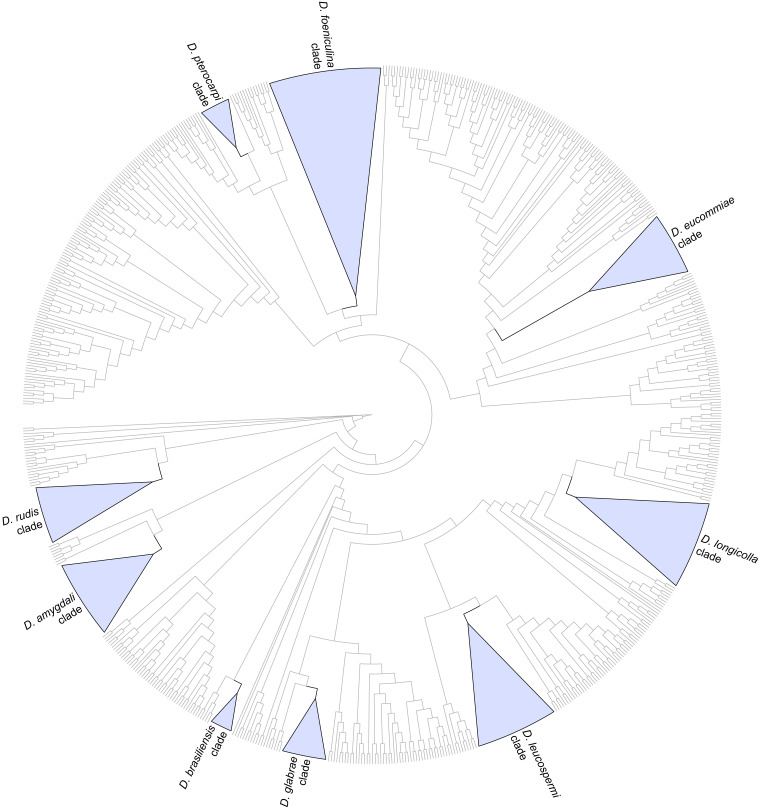
Fig. 1.
Cladogram of the phylogenetic tree generated from a maximum likelihood analysis based on combined ITS, tef1, tub2, cal and his3 sequence data of the isolates from palms examined in this study and all species currently accepted in the genus Diaporthe. A total of 783 strains were included in the combined dataset that comprised 2 790 characters (including gaps) (576 characters for ITS, 507 for tef1, 560 for tub2, 614 for cal and 533 for his3) after alignment and manual adjustment. The analysis was conducted using the GTR+G+I nucleotide substitution model. Taxa names and phylogenetic support values have been removed for simplification purposes. The well-supported (ML-BS ≥ 95 %) clades containing the isolates from palms examined in this study have been collapsed and are highlighted with coloured triangles with their respective branches in black. Cladogram is rooted to Cytospora disciformis (CBS 116827 and CBS 118083).