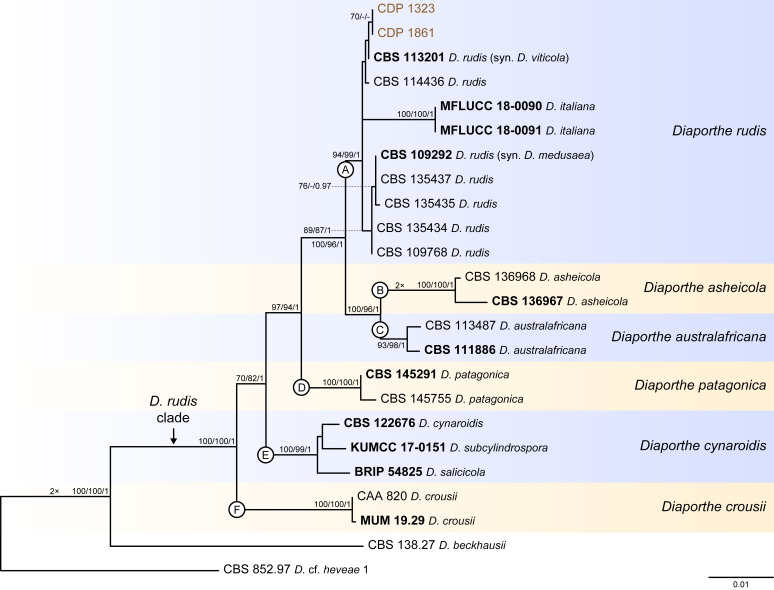
Fig. 10.
Phylogenetic tree generated from a maximum likelihood analysis based on combined ITS, tef1, tub2, cal and his3 sequence data of the Diaporthe rudis clade and closely related species. Bootstrap support values for maximum likelihood, maximum parsimony (ML-BS/MP-BS ≥ 70 %) and Bayesian posterior probabilities (PP ≥ 0.90) are shown at the nodes. Strains with type status are indicated in bold font. The isolates from this study are presented in brown typeface. Species boundaries within the D. rudis clade are delimited with coloured blocks and their respective branches are indicated by lettered circles (A–F). The scale bar represents the expected number of nucleotide changes per site. The tree is rooted to D. cf. heveae 1 (CBS 852.97).