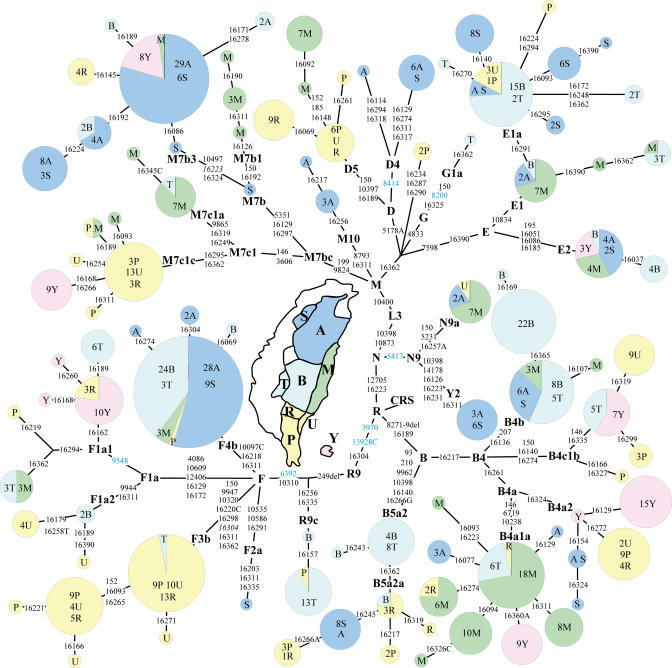
Figure 2. Tree Drawn from a Median-Joining Network of 96 mtDNA Haplotypes Observed in Nine Indigenous Taiwanese Populations.
The tree is based on sequences of HVS-I (16024–16390) and a coding region segment covering 9,793 to 10,899 bps. Haplogroup defining HVS-II mutations were manually added after the generation of the network. Additional coding region mutations, ascertained through complete mtDNA sequencing of an individual of each subclade of the haplogroup defined by the mutation, were used to generate the network and are shown in blue. All nps are numbered according to reference sequence [71]. Mutations in italic indicate back conversions. Nucleotide change is specified only for transversions. Node areas are proportional to haplotype frequencies of the pooled nine tribes. The population codes are as follows: A, Atayal; B, Bunun; M, Amis; P, Paiwan; R, Rukai; S, Saisiat; U, Puyuma; T, Tsou; and Y, Yami. CRS = Cambridge reference sequence [70].