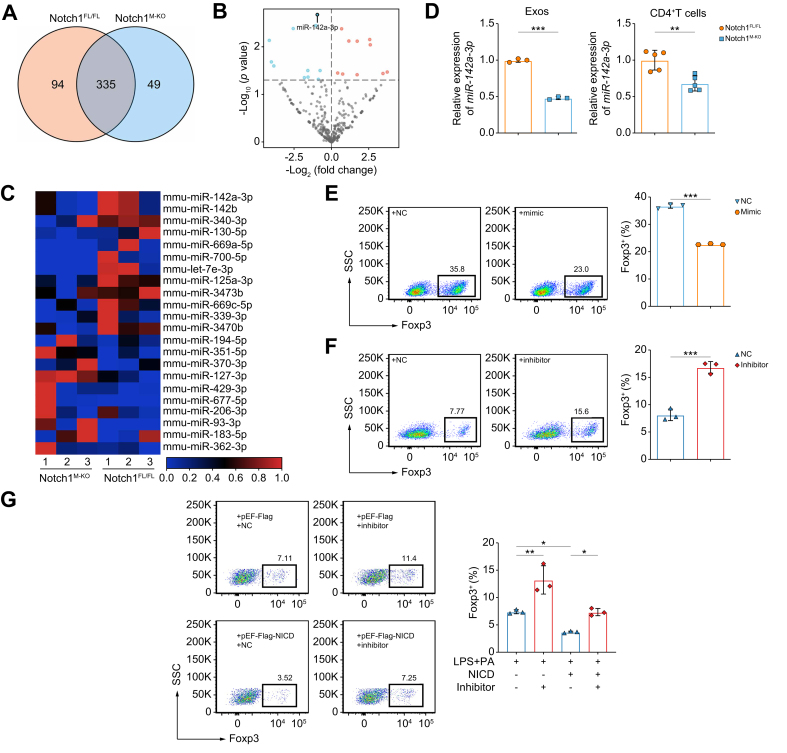
Fig. 6.
Exo-miR-142a-3p from Notch1-activated macrophages impedes Treg differentiation.
BMDMs from Notch1FL/FL and Notch1M-KO mice were stimulated with LPS (100 ng/ml) and PA (250 μM) for 12 h. The miRNA expression profiles in BMDM-Exos were analyzed by miRNA sequencing (three/group). (A) Venn diagram showing miRNAs in both groups. (B) Volcano plot of differentially expressed miRNAs in both groups. (C) Heatmap of miRNAs that differed significantly (p <0.05) between the groups. (D) Validation of the expression of miR-142a-3p in BMDM-Exos (left panel) (three/group) and CD4+ T cells (right panel) (five/group) between the two groups by RT-qPCR analysis. (E) Induction of Foxp3+ Tregs was detected by flow cytometry after naïve CD4+ T cells transfected with NC or miR-142a-3p mimic were co-cultured with BMDMs (three/group). (F) Induction of Foxp3+ Tregs was detected by flow cytometry after naïve CD4+ T cells transfected with NC or miR-142a-3p-inhibitor were co-cultured with BMDMs pretreated with LPS (100 ng/ml) and PA (250 μM) for 12 h (three/group). (G) pEF-Flag-NICD or pEF-Flag transfected BMDMs were stimulated with LPS (100 ng/ml) and PA (250 μM) or PBS for 12 h, respectively, and then co-cultured with naïve CD4+ T cells transfected with NC or miR-142a-3p-inhibitor for 3 days. The induction of Foxp3+ Tregs was analyzed by flow cytometry (three/group). Values represent means ± SD. Statistical analysis was performed by (D–F) 2-tailed unpaired Student t test or (G) one-way ANOVA and Tukey's test: ∗p <0.05, ∗∗p <0.01, ∗∗∗p <0.001. Abbreviations: BMDM, bone marrow-derived macrophage; Exo, exosome; LPS, lipopolysaccharide; NC, negative control; NICD, Notch1 intracellular domain; Notch1FL/FL, floxed Notch1; Notch1M-KO, myeloid-specific Notch1-knockout; PA, palmitic acid; RT-qPCR, quantitative reverse transcription PCR; Treg, regulatory T cell.