Table 2. Best-Fit 11B[p] Chemical Shifts and Fractional Populations before and after 2QFa.
| Fractional Populationb |
Isotropic Chemical Shift (ppm) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
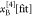 c c
|
11B[3] |
11B[4](mSi) |
11B[3] |
11B[4](mSi) |
|||||||
| Glass | 1pls(2QF) | Ring | Non-ring | 2Si | 3Si | 4Si | Ring | Non-ring | 2Si | 3Si | 4Si |
| K2.0 | 0.660(0.463) | 0.265(0.517) | 0.075(0.020) | 0.157(0.199) | 0.299(0.175) | 0.204(0.089) | 17.95(18.54) | 14.97(17.00) | 1.00(0.97) | –0.37(−0.21) | –1.90(−1.90) |
| 0.779(0.963) | 0.221(0.037) | 0.237(0.430) | 0.453(0.377) | 0.309(0.192) | |||||||
| RbNa2.0 | 0.634(0.435) | 0.300(0.533) | 0.066(0.031) | 0.143(0.223) | 0.315(0.154) | 0.175(0.058) | 17.82(18.26) | 14.56(15.67) | 1.00(1.20) | –0.32(−0.08) | –1.90(−1.70) |
| 0.820(0.944) | 0.180(0.056) | 0.226(0.513) | 0.497(0.355) | 0.277(0.132) | |||||||
| Na2.0 | 0.635(0.472) | 0.276(0.500) | 0.089(0.028) | 0.121(0.154) | 0.284(0.217) | 0.231(0.101) | 17.88(17.90) | 14.59(14.62) | 1.00(1.10) | –0.12(−0.06) | –1.68(−1.70) |
| 0.757(0.947) | 0.243(0.053) | 0.190(0.327) | 0.447(0.459) | 0.364(0.214) | |||||||
| LiNa2.0 | 0.597(0.435) | 0.302(0.529) | 0.101(0.036) | 0.123(0.149) | 0.291(0.222) | 0.183(0.065) | 17.82(18.10) | 14.95(16.10) | 1.05(1.50) | –0.04(0.02) | –1.63(−1.74) |
| 0.749(0.936) | 0.251(0.064) | 0.206(0.342) | 0.488(0.510) | 0.306(0.148) | |||||||
| MgNa2.0 | 0.388(0.277) | 0.409(0.592) | 0.203(0.131) | 0.077(0.089) | 0.193(0.134) | 0.118(0.053) | 17.64(18.09) | 14.44(15.54) | 1.05(1.30) | 0.03(0.10) | –1.62(−1.62) |
| 0.669(0.818) | 0.331(0.182) | 0.198(0.322) | 0.499(0.485) | 0.303(0.193) | |||||||
| K4.0 | 0.713(0.423) | 0.178(0.475) | 0.109(0.102) | 0.057(0.099) | 0.254(0.239) | 0.402(0.086) | 17.50(17.90) | 14.35(14.86) | 1.00(1.30) | –0.58(−0.28) | –2.13(−1.96) |
| 0.621(0.823) | 0.379(0.177) | 0.079(0.233) | 0.356(0.564) | 0.565(0.203) | |||||||
| RbNa4.0 | 0.686(0.402) | 0.206(0.509) | 0.108(0.089) | 0.040(0.073) | 0.289(0.240) | 0.358(0.090) | 17.41(18.44) | 14.06(16.00) | 1.30(1.40) | –0.48(−0.22) | –2.10(−1.90) |
| 0.657(0.852) | 0.343(0.148) | 0.058(0.181) | 0.421(0.596) | 0.522(0.222) | |||||||
| Na4.0 | 0.650(0.414) | 0.214(0.525) | 0.135(0.061) | 0.050(0.090) | 0.283(0.237) | 0.318(0.086) | 17.46(18.15) | 14.13(15.88) | 1.07(1.30) | –0.40(−0.26) | –2.02(−2.02) |
| 0.613(0.895) | 0.387(0.105) | 0.076(0.217) | 0.435(0.574) | 0.489(0.208) | |||||||
| MgNa4.0 | 0.338(0.247) | 0.360(0.629) | 0.303(0.124) | 0.042(0.048) | 0.182(0.138) | 0.113(0.061) | 17.52(18.30) | 13.83(14.54) | 1.13(1.37) | –0.17(−0.03) | –1.96(−1.80) |
| 0.543(0.835) | 0.457(0.165) | 0.125(0.196) | 0.540(0.556) | 0.334(0.248) | |||||||
Fractional populations and isotropic 11B chemical shifts obtained by deconvoluting the 11B MAS NMR spectrum obtained directly by single-pulse excitation (“1pls”) and the projection along the 1Q dimension of the 2Q–1Q correlation 2D NMR spectrum (“2QF”; values within parentheses). Data uncertainties are listed in Table S2.
Fractional populations
normalized
to a unity sum over all B[3] and B[4] species
in the glass (top row for each glass). The row beneath lists the corresponding
data normalized to a unity sum within each set of B[3] and
B[4] populations:  and
and 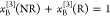 , where “R” and “NR”
represent “ring” and “non-ring” sites,
respectively.
, where “R” and “NR”
represent “ring” and “non-ring” sites,
respectively.
Net fractional
population of the
best-fit {x[4]B(mSi)} set: 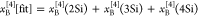 . The minor discrepancy to its
. The minor discrepancy to its 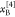 counterpart (“1pls”) listed
in Table 1 stems partially
from uncertainties in the spectral deconvolutions but primarily from
the centerband ST 11B[4] resonance intensity,
which was not accounted for by the spectral deconvolution and leading
to an overestimation of
counterpart (“1pls”) listed
in Table 1 stems partially
from uncertainties in the spectral deconvolutions but primarily from
the centerband ST 11B[4] resonance intensity,
which was not accounted for by the spectral deconvolution and leading
to an overestimation of 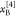 by ≈ 0.03.
by ≈ 0.03.
