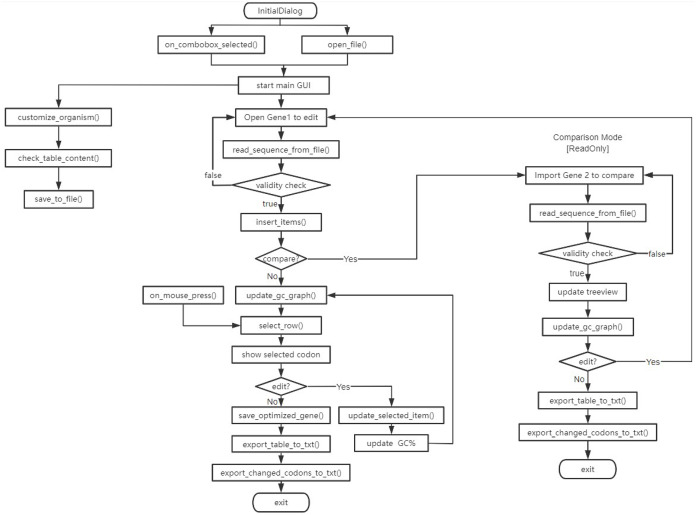
Figure 1. Flowchart of program.
InitialDialog: Launch the initial screen of the program; ‘start main GUI’: launch the program and open the main interface; customize_organism(): customize the codon table usage of host species; check_table_content(): check whether the content filled in meets the requirements; save_to_file(): save the codon table usage of host species to a txt file; ‘Open Gene 1 to edit’: open a FASTA gene file; read_sequence_from_file(): read a sequence file; ‘validity check’: check the validity of the sequence file; insert_itmes(): insert an item to the Treeview; update_gc_graph(): plot the GC content of the gene. update_selected_item(): change the codon of the selected amino acid; save_optimized_gene(): save optimized gene sequence to a FASTA file; export_table_to_txt(): export the Treeview table to a txt file; export_changed_codons_to_txt(): export the changed codons to a txt file; ‘Import Gene 2 to compare’: import a fasta or fa gene file, which codes the same protein as ‘Open Gene 1 to edit’. The program’s mode will become read-only.