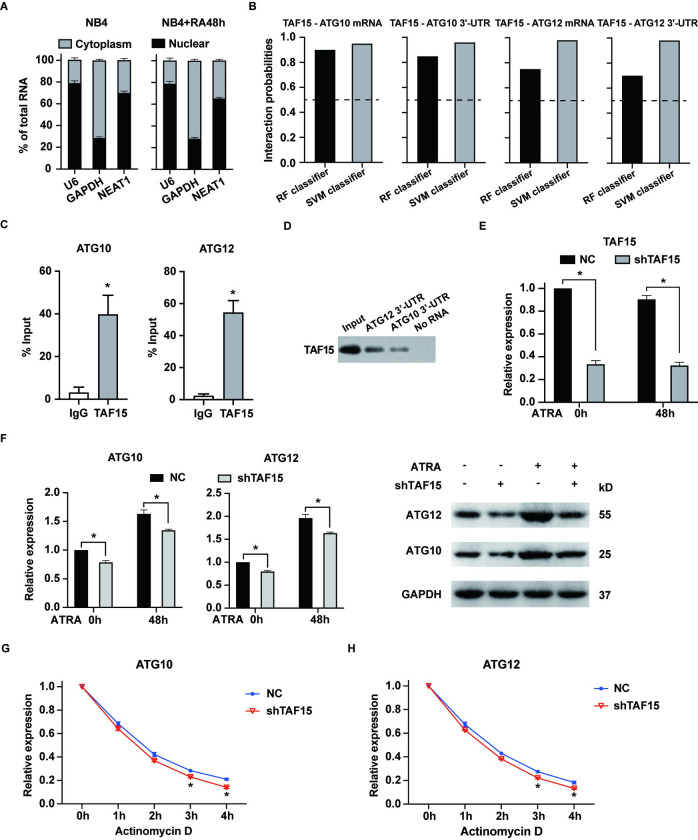
Fig 3. TAF15 binds to and stabilizes ATG10 and ATG12 mRNA in NB4 cells.
(A) The expression of NEAT1 in the cytoplasm and nucleus of NB4 cells, both untreated and treated with ATRA, was determined by RT-qPCR. GAPDH served as a marker for the cytoplasm, and U6 as a marker for the nucleus. (B) The likelihood of TAF15 binding to the mRNA/3’-UTR regions of ATG10 and ATG12 were predicted by the RNA-Protein interaction prediction (RPISeq) website. “RF classifier” means random forest classifier and “SVM classifier” means support vector machine classifier. A prediction probability >0.5 was considered “positive”, indicating a probable interaction between the RNA and protein. (C) RIP experiments were conducted in ATRA-treated NB4 cells to confirm the interaction between TAF15 and the mRNA of ATG10 and ATG12. (D) Biotinylated probes of the 3’-UTR regions of ATG10 and ATG12 were employed to pull down TAF15 from cell lysates. The presence of TAF15 in the precipitates was detected using western blot assay. (E) NB4 cells were transfected with shRNA specifically targeting TAF15 (shTAF15) or negative control shRNA (NC). TAF15 expression was detected by RT-qPCR in these cells both before and after treatment with 1 μM ATRA for 48 hours. (F) The RNA and protein levels of ATG10 and ATG12 were measured in TAF15-silenced NB4 cells before and after ATRA treatment for 48 hours. (G-H) NB4 cells with or without TAF15 knockdown were treated with 1 μM ATRA for 48 hours, then the cells were further treated with Actinomycin D to inhibit new RNA synthesis. The expression levels of ATG10 and ATG12 mRNA were determined every hour by RT-qPCR. The data represent the mean ± S.E.M. from three replicates. * indicates p<0.05.